Chromatin replication and parental histone allocation
Genome Instability & Disease 2, 51–58(2021)
Abstract
The propagation of epigenetic information across cell divisions is crucial to the maintenance of cell identity. During DNA replication, both parental and newly synthesized histones are deposited onto the replicating DNA, where they assemble into nucleosomes. Parental histones bear a variety of post-translational modifications, which constitute crucial epigenetic information, and the pattern of parental and newly synthesized histone allocation is crucial for the determination and maintenance of cell fate in multicellular organisms. The DNA replication-coupled nucleosome assembly process is regulated by multitude of histone chaperones, and recent breakthroughs in next-generation sequencing and super-resolution imaging methods have provided novel insights into the underlying mechanisms. Here, we review recent findings concerning parental histone deposition onto replicating DNA strands and the segregation of these to symmetrically or asymmetrically dividing daughter cells. Furthermore, we discuss the role of the allocation patterns of parental and newly synthesized histones in the determination of cell fate.
Introduction
In eukaryotes, genetic information is reliably transferred from parental cells to daughter cells during cell division through the process of DNA replication. Along with chromatin assembly, DNA replication is also heavily involved in the inheritance of epigenetic information (Alabert et al. 2017b; Allis and Jenuwein 2016; Miller and Costa 2017). Histone and its post-translational modifications (PTMs) represent one of the crucial epigenetic components that help specify cell fates. Trimethylated histone H3 Lys9 (H3K9me3), a mark that is commonly associated with constitutive heterochromatin regions in cells, acts as a key chromatin barrier to cell fate changes (Peters et al. 2001; Probst et al. 2009). The histone chaperone CAF-1 is found to maintain somatic cell identities by maintaining chromatin landscape, thus maintaining the transcriptional program of a specific cell type. Reducing the levels of CAF-1 accelerated the reprogramming efficiency of induced pluripotent stem cells from various differentiated cell types (Cheloufi et al. 2015; Ishiuchi et al. 2015). Chromatin factors contribute to the chromatin landscape during cell division and overall organism development.
In most tissues, stem cells have to ensure a balance between the generation of differentiated cells, and the preservation of a stem cell population (Knoblich 2008). This can be achieved through symmetric cell divisions by self-renewing or differentiating, or asymmetric cell divisions through which two daughter cells are produced, each with their own distinct cell fate-daughter stem cell versus daughter differentiating cell. Balance between these different divisions is critical for homeostasis and organism development (Morrison and Kimble 2006). Given the crucial roles of epigenetic mechanisms play in regulating chromatin patterns and gene expression landscape that defines distinct cell identities, nucleosome reassembly, a key epigenetic determinant, may serve as an important mechanism in the determination of cell fate (Escobar et al. 2019; Schlissel and Rine 2019).
Nucleosomes are the basic units of chromatin. Each nucleosome is composed of two H2A–H2B histone dimers and one H3–H4 histone tetramer, and is wrapped with 147 DNA base pairs (~ 1.6 turns). Linker DNA connects the nucleosomes like “beads on a string”, forming the primary structure of chromatin (McGinty and Tan 2015). DNA replication is a disruptive process that variety of histone chaperones accompany replication forks and help to re-assemble chromatin after the forks (Stewart-Morgan et al. 2020). A number of histone chaperones accompany these replication forks and help to re-assemble the chromatin after replication. These histones include both newly synthesized and parental histones, with the parental histones bearing crucial epigenetic information in the form of post-translational modifications (Serra-Cardona and Zhang 2018). Next-generation sequencing (NGS), super-resolution images of chromatin fibers, and dual-color switching system techniques have revealed that these parental histones show different allocation patterns to newly synthesized histones (Gan et al. 2018; Ma et al. 2020; Petryk et al. 2018; Tran et al. 2012; Wooten et al. 2019; Yu et al. 2018). Disruption to these deposition patterns is likely to result in aberrant cell fate and differentiation (Xie et al. 2015). In this review, we discuss recent progress in understanding the mechanisms underlying parental histone, allocation, and the role of this process in cell fate and differentiation.
Recycling of parental histone and recruitment of newly synthesized histone during the process of replication-coupled nucleosome assembly
During DNA replication, chromatin proteins, and RNA are transiently disassociated from the DNA and the nucleosomes are disassembled. Inactive MCM2-7 double hexamers are loaded around the double-stranded DNA (dsDNA) by the origin recognition complex (ORC) in G1 phase. Subsequently, in S phase, cyclin-dependent kinases (CDKs) and Dbf4-dependent kinase (DDK) split the MCM2-7 double hexamer and recruit Cdc45 protein and the GINS (Go–Ichi–Ni–San) complex. Following this, two active Cdc45-MCM helicase-GINS (CMG) helicases initiate a pair of bidirectional replication forks. Each CMG helicase travels along the template of the leading strand, unwinding the parental dsDNA template (Fu et al. 2011; Yu et al. 2014). Next, DNA polymerase synthesizes DNA in the 5′-to-3′ direction, with the leading strand being synthesized rapidly and continuously by DNA polymerase epsilon (Pol ε), while the lagging strand is synthesized discontinuously by DNA polymerases alpha (Pol α) and delta (Pol δ). The lagging strand is synthesized in interspersed segments of RNA primers and new DNA fragments. The RNA primers are subsequently displaced by short Okazaki fragments, removed by FEN1, and ligated by DNA ligase 1 to complete the double-stranded daughter chromatid (Burgers and Kunkel 2017).
The DNA replication machinery contains factors involved in chromatin duplication as well as DNA replication (Bell and Dutta 2002; DePamphilis 2000). Immediately after DNA replication, chromatin components are re-assembled behind the replication fork on the newly replicated DNA (Stewart-Morgan et al. 2020). Parental histones, which are inherited from parental strands, are recycled and deposited onto replicating DNA strands, while newly synthesized histones are recruited de novo and deposited to restore histone levels. Nascent chromatin contains both parental and newly synthesized histones, and parental histone recycling and new histone recruitment are together referred to as replication-coupled nucleosome assembly (Serra-Cardona and Zhang 2018) (Fig. 1a). The two histone types can be differentiated by their distinct post-translational modifications. For instance, acetylation at lysine 5 (H4K5ac) and deacetylation at histone H4 lysines 5 and 12 (H4K5ac2 and H4K12ac2) are markers of newly synthesized histones in eukaryotes; as is acetylation at histone H3 lysine 56 (H3K56ac) in yeast. Di-methylation at lysine 20 (H4K20me2) and tri-methylation of histone H3 lysine 4 (H3K4me3) are markers of parental histones (Annunziato 2015; Loyola et al. 2006; Scharf et al. 2009). Histone H3–H4 tetramers are relatively stable parts of the nucleosome structure, while Histone H2A–H2B dimers can rapidly exchange with cytoplasmic H2A–H2B during DNA replication (Xu et al. 2010). Therefore, parental H3–H4 histones are likely recycled as intact tetramers, while H2A and H2B histones are incorporated as dimers (Alabert et al. 2015; Xu et al. 2010). This means that each parental histone H3–H4 tetramer can only be transferred to one strand, while parental H2A–H2B dimers can be re-assembled into nucleosomes on both the leading and the lagging strands. Parental histones and new histones are deposited almost in a 1:1 ratio in human HeLa cells, mouse embryonic stem cells, and budding yeast (Alabert et al. 2015; Petryk et al. 2018; Yu et al. 2018). The newly synthesized and parental histones were tracked by quantitative mass spectrometry immediately after their deposition behind the replication fork using nascent chromatin capture. In nascent chromatin capture, biotin-dUTP labeling of newly replicated DNA and cross-linking of associated proteins allow the isolation of nascent chromatin. Then, nascent chromatin capture was combined with SILAC (stable isotope labeling with amino acids in cell culture) to differentiate newly synthesized histones (heavy) and old recycled histones (light). The nascent chromatin purified by nascent chromatin capture contained approximately equal amounts of new and old histones (Alabert et al. 2015). In other studies, the newly synthesized and parental histones were differentiated by distinct post-translational modifications, combined with nascent chromatin capture and genome-wide mapping, and the new and old histones are deposited equally in leading and lagging strands (Gan et al. 2018; Petryk et al. 2018; Yu et al. 2018).
Fig. 1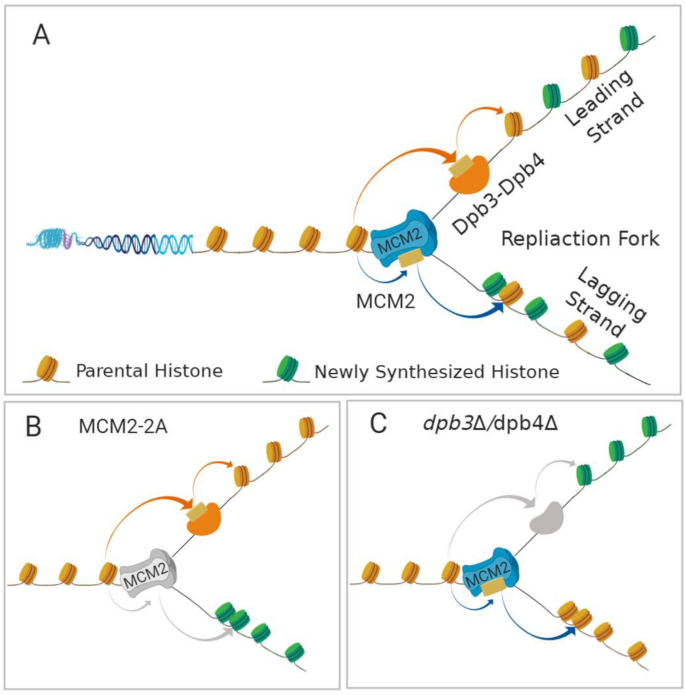
Parental histone recycling pathway during DNA replication. a DNA replication-coupled nucleosome assembly at the replication fork. MCM2, a subunit of the MCM2-7 replicative helicase traveling along leading strand, transfers and deposits parental histone to the lagging strand. Dpb3–Dpb4, two auxiliary and non-essential subunits of leading-strand DNA polymerase ε, transfer and deposit parental histone to the leading strand. b Mutations of MCM2 that disrupt the chaperone activities impair parental histone recycling to the lagging strand. The role of MCM2 chaperone activity is conserved between budding yeast and mouse. c Deletion of Dpb3 or Dpb4 that impairs parental H3–H4 recycling to the leading strand. The roles of Dpb3 and Dpb4 in histone partitioning have only been reported in budding yeast
Full size imageHistones are modified by a variety of PTMs, including phosphorylation, acetylation, methylation, ubiquitination, and ADP ribosylation (Allis and Jenuwein 2016; Stillman 2018). These histone PTMs have a profound impact on the structure of chromatin and, subsequently, on gene expression (Bannister and Kouzarides 2011). In most species, repressive chromatin is generally enriched in H3K9 and H3K27 tri-methylation (H3K9me3 and H3K27me3), while H3K4me3 is enriched at promoter regions and associated with more accessible chromatin (Black et al. 2012). Parental H3–H4 histones preserve their PTMs during DNA replication and chromatin reassembly (Alabert and Groth 2012; Alabert et al. 2017a). This represents epigenetic information, and may passed on to daughter cells during cell division via parental H3-H4 histones, to maintain cellular identity or confer specific cellular differentiation.
Using advanced analytical techniques to investigate parental histone allocation and distribution
The molecular mechanisms governing the deposition of parental histone H3–H4 tetramers onto replicating DNA strands remain to be fully understood, but the use of advanced analytical techniques has rapidly progressed our understanding of histone allocation (Liu et al. 2017; Serra-Cardona and Zhang 2018; Wooten et al. 2020; Zhang et al. 2017). Major breakthroughs have been made using high-throughput next-generation sequencing (NGS), super-resolution images of chromatin fibers, and dual-color switching system techniques, and these are summarized below.
Characterization of parental histone allocation onto leading and lagging strands using NGS-based methods
NGS-based techniques have been used to study replication-coupled histone inheritance patterns in yeast and mouse embryonic stem cells. Enrichment and Sequencing of Protein-Associated Nascent DNA (eSPAN) (Gan et al. 2018; Yu et al. 2018) and Sister Chromatids After Replication by DNA sequencing (SCAR-seq) (Petryk et al. 2018) are two similar NGS-based techniques that characterize the distribution of parental and new histones at BrdU- or EdU-labeled newly synthesized DNA strands of the replication fork. The eSPAN method uses genome-wide mapping, and has been used to track the distribution of histones at newly replicated Watson and Crick DNA strands in both budding yeast (Saccharomyces cerevisiae) (Yu et al. 2018) and mouse embryonic stem cells (Li et al. 2020). The SCAR-seq method maps Okazaki fragments to determine replication fork directionality, and then maps parental and newly synthesized histone deposition across the genome. This technique has also been applied in mouse embryonic stem cells (Petryk et al. 2018). These methods found that parental histones were deposited to the leading and lagging strands almost equally in mouse embryonic cells and budding yeast, with new histones being deposited in the remaining space across the replicating DNA strands (Petryk et al. 2018; Yu et al. 2018). In contrast, in budding yeast, the eSPAN method detected a slight lagging-strand preference in the deposition of parental histone H3–H4 tetramers (Yu et al. 2018), while SCAR-seq found a slight bias toward the leading strand in the distribution of parental histones (Petryk et al. 2018).
These studies have revealed that parental histone allocation is carefully orchestrated by specific histone chaperones, which capture histones displaced from the parental nucleosome and transfer them to the leading and lagging strands (Table 1). MCM2, a subunit of MCM2-7 replicative helicase, preferentially binds histone H3–H4 and has been found to facilitate the transfer of parental H3–H4 tetramers to the lagging strands (Petryk et al. 2018). Further studies have revealed that MCM2, along with the adaptor protein Ctf4 and DNA polymerase α (Pol α), which synthesizes short RNA primers, forms the MCM2–Ctf4–Pol α axis within the replisome (Gan et al. 2018; Li et al. 2020) (Fig. 1a). Loss of MCM2 or Pol α chaperone activity through mutations to their histone-binding domains results in a dramatic loss of symmetric histone deposition, with parental histones being predominantly deposited on the leading strand and newly synthesized histones on the lagging strand (Gan et al. 2018; Li et al. 2020; Petryk et al. 2018) (Fig. 1b). In addition, Dpb3 and Dpb4, two auxiliary, non-essential subunits of leading-strand DNA polymerase ε (Pol ε), also facilitate the deposition of parental histone H3–H4 tetramers to the leading strands. Deletion of either Dpb3 or Dpb4 compromises this leading-strand preference (Yu et al. 2018) (Fig. 1c). MCM2, Pol α, Dpb3, and Dpb4 function together at the DNA replication fork to control the allocation of parental histones, along with their preserved PTMs, onto replicating leading and lagging strands and so to daughter cells Fig. 2.
Table 1 Details of histone chaperone that transferring parental histone during DNA replication
| Histone-binding proteins | Symbol | Complex | Model | Technique | Parental histone strand bias | References |
|---|---|---|---|---|---|---|
| Minichromosome maintenance complex component 2 | MCM2 | MCM2-7 complex, replicative helicase | mESC S. cerevisiae | eSPAN SCAR-Seq | Lagging | (Gan et al. 2018; Petryk et al. 2018) |
| DNA polymerase alpha 1, catalytic subunit | POLA1 | Catalytic subunit of Pol α primase | mESC S. cerevisiae | eSPAN | Lagging | (Gan et al. 2018; Li et al. 2020) |
| DNA polymerase epsilon Dpb3, noncatalytic subunit | Dpb3 | Subunit of Pol ε | S. cerevisiae | eSPAN | Leading | (Yu et al. 2018) |
| DNA polymerase epsilon Dpb4, noncatalytic subunit | Dpb4 | Subunit of Pol ε | S. cerevisiae | eSPAN | Leading | (Yu et al. 2018) |
mESC mouse embryonic stem cell, eSPAN enrichment and sequencing of protein-associated nascent DNA, SCAR-seq sister chromatids after replication by DNA sequencing
Full size tableFig. 2
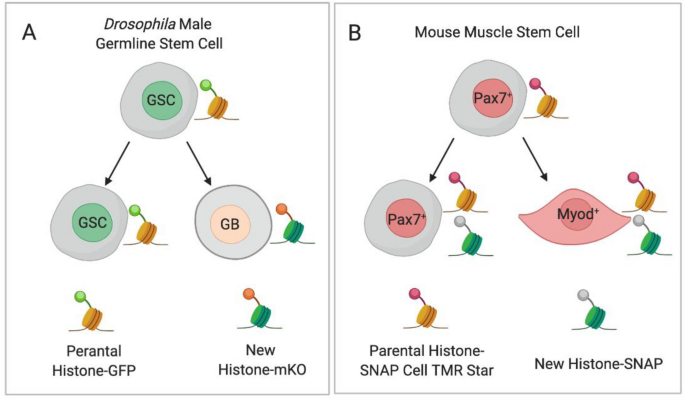
Symmetric and asymmetric allocation of parental histones in asymmetric dividing stem cells. a Parental histones are labeled with GFP and new histone with monomeric Kusabira-Orange (m-KO) by the temporally heat shock-controlled dual-color switching system in the Drosophila germline stem cells. The parental histone is segregated to the self-renewed germline stem cell, whereas new histone is enriched toward the differentiating daughter cell during asymmetric divisions of Drosophila male germline stem cells. b Parental histones are incubated with fluorescent substrate SNAP Cell TMR Star before DNA replication, while the newly synthesized histones are SNAP fluorescent negative in H3-SNAP transgene mouse muscle stem cells. The parental histones show a symmetric distribution during asymmetric dividing of muscle stem cells. GSC Drosophila germline stem cell, GB Drosophila gonialblast, Pax7+ Pax7 positive muscle stem cells, Myod+ Myod positive differentiated muscle cell
Full size imageCharacterization of histone incorporation patterns using super-resolution imaging of chromatin fibers
Super-resolution imaging of chromatin fibers method was used to directly visualize histone deposition patterns at replication forks in asymmetrically dividing Drosophila male germ cells (Wooten et al. 2019) and mouse embryonic stem cells (Ma et al. 2020). The parental histones were labeled by H3K27me3 or H4K20me2/3, while lagging strand was labeled by eGFP-RpA-70 (replication protein-A) or PCNA-eGFP respectively (Ma et al. 2020; Wooten et al. 2019). The parental histones were found to enrich toward the eGFP-RpA-70 or PCNA-eGFP negative leading strands. Further studies reveal that parental H3-eGFP and new H3-monomeric Kusabira-Orange deposited on distinct strands (Wooten et al. 2019). Thus, parental histones were deposited on the leading strand at the replication fork, while newly synthesized histones were enriched on the lagging strand (Ma et al. 2020; Wooten et al. 2019).
Dual-color switching systems and pulse-chase labeling approaches reveal parental histone distribution in daughter cells
Dual-color switching systems are also useful tools for studying the distribution of parental and newly synthesized histones (Table 2). For example, a temporally heat shock-controlled dual-color switching system was used to precisely label parental histones with GFP and newly synthesized histones with monomeric Kusabira-Orange in the germline of Drosophila melanogaster germline cells. This revealed that, during asymmetric divisions of these germline stem cells, parental H3 and H4 histones were selectively segregated to the self-renewed germline stem cell, while newly synthesized histone H3 and H4 was enriched toward the differentiating daughter cells (Fig. 2a) (Tran et al. 2012, 2013). This asymmetric pattern is distinct from the symmetrical H3 inheritance found in symmetrically dividing germline stem cells. In addition, H2A and H2B have been found to be symmetrically inherited during asymmetric divisions of germline stem cells (Wooten et al. 2019). Subsequently, a photoconvertible dual-color switching system was used to label parental histones with red Dendra2 and newly synthesized H3 with green Dendra2 in mouse embryonic stem cells. During induced asymmetric divisions of these embryonic stem cells, parental histone H3 and H4 was selectively segregated to the daughter stem cells, while newly synthesized histone H3 and H4 was enriched toward the epiblast stem daughter cell (Ma et al. 2020). This pattern of histone distribution was similar to that found in the Drosophila male germline stem cells under asymmetric cell division. The asymmetric H3 and H4 inheritance mode may serve as a more general mechanism during the asymmetric cell division of stem cell in Drosophila and mouse embryonic stem cell, and in germline stem cell and embryonic stem cell.
Table 2 Parental histone allocation during stem cell asymmetric dividing
| Cell type | Model | Technique | Allocation pattern of parental histone | Parental histone cell bias | References |
|---|---|---|---|---|---|
| Germline stem cell | Drosophila | Temporally heat shock-controlled DCSS, SRCF | Asymmetry | Daughter stem cell | (Tran et al. 2012; Wooten et al. 2019) |
| Embryonic stem cell | Mouse | Photoconvertible DCSS, SRCF | Asymmetry | Unknown | (Ma et al. 2020) |
| Muscle stem cell | Mouse | SNAP tag pulse-chase labeling approach | Symmetry | No bias | (Evano et al. 2020) |
SRCF super-resolution of chromatin fibers, DCSS dual-color switching system
Full size table
Super-resolution imaging of chromatin fibers revealed that, during asymmetric division in Drosophila male germline stem cells, parental histones were preferentially deposited onto the leading strand, while new histones were deposited onto the lagging strand. However, as the replication forks proceed in a bidirectional manner, the leading strand and its parental histones alternate with the lagging strand and its new histones in a sister chromatid. Using EdU and BrdU incorporation chromatin fiber technology, the replication forks of chromatin fibers isolated from Drosophila testes were found to move in unidirectional or asymmetric bidirectional patterns. These fork movement patterns allowed most of the parental or the new histones to be deposited onto a sister chromatid (Wooten et al. 2019). Subsequently, centromere protein-A (CENP-A), a histone H3 variant deposited in the centromere, was also found to be asymmetrically segregated in asymmetrically dividing Drosophila male germline stem cells. Daughter germline stem cells contained more CENP-A than differentiating daughter cells, and is asymmetrically dividing germline stem cells in prometaphase, and the centromere with more CENP-A was found to attach to the daughter stem cell side with microtubules (Ranjan et al. 2019; Xie et al. 2015). Unidirectional and asymmetric bidirectional forks combined with asymmetric sister chromatid recognition and segregation lead to the asymmetric allocation of parental and newly synthesized histone allocation into two daughter cells, each with a different cell fate.
A pulse-chase labeling approach using SNAP tags has also been utilized to track the deposition of parental and newly synthesized histones (Table 2). H3-SNAP transgene mouse muscle stem cells were incubated with a fluorescent SNAP substrate to label parental histones, while the newly synthesized histones were free of SNAP fluorescence. The symmetric and asymmetric division of the muscle stem cells could then be traced at a single-cell resolution. However, unlike in Drosophila germline stem cells and mouse embryonic stem cells, the parental histones were evenly distributed between the muscle daughter cells (Fig. 2b) (Evano et al. 2020). The difference observed between Drosophila germline stem cells and mouse embryonic stem cells may be due to the differences in the approaches used, or mechanistic differences between germline stem cells and embryonic stem cells versus somatic stem cells.
Parental histone allocation affects cell maintenance and fate
The disruption of asymmetric parental histone inheritance in Drosophila germline stem cells has been found to lead to both early germline tumors and germline stem cell loss (Wooten et al. 2020; Xie et al. 2015, 2017). The CAF-1 protein complex is an evolutionarily conserved histone chaperone that deposits histone H3–H4 proteins onto replicating DNA, and is required to generate a bilateral asymmetry in the C. elegans nervous system. Loss of CAF-1 chaperone activity via mutations in its histone-binding domain leads to defects in bilateral asymmetry in the nervous system, as the asymmetrical allocation of parental and new histones is required for the development of asymmetry in the C. elegans nervous system (Nakano et al. 2011). However, limited information concerning the role of parental histone allocation in other organisms is available at present.
Cell reprogramming, trans-differentiation, and pluripotency maintenance studies have found that the histone chaperone CAF-1 stabilizes the structure of chromatin by regulating nucleosome assembly and the formation of heterochromatin (Cheloufi et al. 2015; Cheloufi and Hochedlinger 2017). Furthermore, the position of parental histones on the replicating DNA is highly conserved. Parental histones are precisely deposited on their original positions in the repressed domains following DNA replication, but not in the transcriptionally activated sites (Escobar et al. 2019; Reverón-Gómez et al. 2018; Schlissel and Rine 2019). Notably, parental histone PTM levels are diluted after DNA replication, but the majority of these can be restored within a cell cycle through the modification of new histones (Alabert et al. 2015; Reverón-Gómez et al. 2018). In certain cases, a substantial delay relative to DNA replication was observed at the H3K27me3 or H3K9me3 domains, leading to the removal of PTMs in long-term cell culture in human somatic cells (Alabert et al. 2015). However, in mouse embryonic stem cells, the H3K27me3 landscape was found to be established de novo in the absence of parental H3K27me3 (Hojfeldt et al. 2018). Disruption of parental histone deposition patterns will alter their original localization on the replicating DNA strands, which in turn will alter chromatin status and gene expression, subsequently affecting the fate of daughter cells. However, the distinct restoration patterns of histone PTMs suggest that the epigenetic landscape may be maintained by an epigenetic buffer system (Huang et al. 2013). This buffer system can tolerate a certain degree of fluctuation when epigenetic modification occurs. A number of alterations to histone PTMs at certain sites can be tolerated with little impact on gene expression, meaning that disruptions to histone deposition patterns can accumulate over many generations of DNA replication.
Summary
DNA replication and nucleosome assembly are closely associated processes that share certain key proteins, including the MCM2-Ctf4-Pol α axis and the histone H3–H4 chaperone Dpb3/Dpb4. The finding that MCM2 and Dpb3/Dpb4, both classical DNA replication machinery components, are involved in the transfer and allocation of parental histones has broadened our understanding of this important cellular process and its role in epigenetic inheritance. Different parental histone allocation patterns have been reported in Drosophila and mouse embryonic stem cells compared with mouse muscle stem cells, using dual-color switching methods and SNAP tag (Evano et al. 2020; Ma et al. 2020; Tran et al. 2012; Wooten et al. 2019).
In the future, genome-editing technologies such as CRISPR-Cas9 may provide further avenues for study: for example, interruption of MCM2 or Dpb3/Dpb4 chaperone activity would allow for further study of parental histone allocation patterns in symmetric and asymmetric cell division. In addition, the loss of MCM2 or Dpb3/Dpb4 chaperone activity may provide novel insights into the roles of histone chaperones in cell differentiation and development in multicellular organisms.
Major breakthroughs regarding DNA replication and histone allocation have been contributed by recent studies, but our understanding of the replication-coupled nucleosome assembly process remains limited. This is largely due to limitations in methods for screening new molecules. With the help of new technologies, such as single-cell analysis, proximity labeling, super-resolution microscopy, and Nanopore sequencing, we expect to gain further insights into the process of epigenetic inheritance-associated chromatin replication.
References
Alabert, C., Barth, T. K., Reveron-Gomez, N., Sidoli, S., Schmidt, A., Jensen, O. N., et al. (2015). Two distinct modes for propagation of histone PTMs across the cell cycle. Genes & Development., 29, 585–590.
Alabert, C., & Groth, A. (2012). Chromatin replication and epigenome maintenance. Nature Reviews. Molecular Cell Biology., 13, 153–167.
Alabert, C., Jasencakova, Z., & Groth, A. (2017a). Chromatin replication and histone dynamics. In H. Masai & M. Foiani (Eds.), DNA replication: from old principles to new discoveries (pp. 311–333). Singapore: Springer Singapore.
Alabert, C., Jasencakova, Z., & Groth, A. (2017b). Chromatin replication and histone dynamics. Advances in Experimental Medicine and Biology, 1042, 311–333.
Allis, C. D., & Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nature Reviews. Genetics., 17, 487–500.
Annunziato, A. T. (2015). The fork in the road: histone partitioning during DNA replication. Genes (Basel)., 6, 353–371.
Bannister, A. J., & Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Research, 21, 381–395.
Bell, S. P., & Dutta, A. (2002). DNA replication in eukaryotic cells. Annual Review of Biochemistry., 71, 333–374.
Black, J. C., Van Rechem, C., & Whetstine, J. R. (2012). Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular Cell, 48, 491–507.
Burgers, P. M. J., & Kunkel, T. A. (2017). Eukaryotic DNA replication fork. Annual Review of Biochemistry., 86, 417–438.
Cheloufi, S., Elling, U., Hopfgartner, B., Jung, Y. L., Murn, J., Ninova, M., et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature, 528, 218–224.
Cheloufi, S., & Hochedlinger, K. (2017). Emerging roles of the histone chaperone CAF-1 in cellular plasticity. Current Opinion in Genetics & Development, 46, 83–94.
DePamphilis, M. L. (2000). Review: nuclear structure and DNA replication. Journal of Structural Biology, 129, 186–197.
Escobar, T. M., Oksuz, O., Saldana-Meyer, R., Descostes, N., Bonasio, R., & Reinberg, D. (2019). Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA replication. Cell, 179(953–963), e911.
Evano, B., Khalilian, S., Le Carrou, G., Almouzni, G., & Tajbakhsh, S. (2020). Dynamics of asymmetric and symmetric divisions of muscle stem cells in vivo and on artificial niches. Cell Reports., 30(3195–3206), e3197.
Fu, Y. V., Yardimci, H., Long, D. T., Guainazzi, A., Bermudez, V. P., Hurwitz, J., et al. (2011). Selective Bypass of a Lagging Strand Roadblock by the Eukaryotic Replicative DNA Helicase. Cell, 146(6), 931–941.
Gan, H., Serra-Cardona, A., Hua, X., Zhou, H., Labib, K., Yu, C., & Zhang, Z. (2018). The Mcm2-Ctf4-polalpha axis facilitates parental histone H3–H4 transfer to lagging strands. Molecular Cell, 72(140–151), e143.
Hojfeldt, J. W., Laugesen, A., Willumsen, B. M., Damhofer, H., Hedehus, L., Tvardovskiy, A., et al. (2018). Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nature Structural & Molecular Biology., 25, 225–232.
Huang, C., Xu, M., & Zhu, B. (2013). Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks? Philosophical Transactions of the Royal Society of London. Series B, Biological sciences, 368, 20110332.
Ishiuchi, T., Enriquez-Gasca, R., Mizutani, E., Bošković, A., Ziegler-Birling, C., Rodriguez-Terrones, D., et al. (2015). Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nature Structural & Molecular Biology., 22, 662–671.
Knoblich, J. A. (2008). Mechanisms of asymmetric stem cell division. Cell, 132, 583–597.
Li, Z., Hua, X., Serra-Cardona, A., Xu, X., Gan, S., Zhou, H., et al. (2020). DNA polymerase α interacts with H3–H4 and facilitates the transfer of parental histones to lagging strands. Science Advances, 6, 5820.
Liu, S., Xu, Z., Leng, H., Zheng, P., Yang, J., Chen, K., et al. (2017). RPA binds histone H3–H4 and functions in DNA replication–coupled nucleosome assembly. Science (New York, N.Y.)., 355, 415–420.
Loyola, A., Bonaldi, T., Roche, D., Imhof, A., & Almouzni, G. (2006). PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Molecular Cell, 24, 309–316.
Ma, B., Trieu, T. J., Cheng, J., Zhou, S., Tang, Q., Xie, J., et al. (2020). Differential histone distribution patterns in induced asymmetrically dividing mouse embryonic stem cells. Cell Reports., 32, 108003.
McGinty, R. K., & Tan, S. (2015). Nucleosome structure and function. Chemical Reviews, 115, 2255–2273.
Miller, T. C., & Costa, A. (2017). The architecture and function of the chromatin replication machinery. Current Opinion in Structural Biology, 47, 9–16.
Morrison, S. J., & Kimble, J. (2006). Asymmetric and symmetric stem-cell divisions in development and cancer. Nature, 441, 1068–1074.
Nakano, S., Stillman, B., & Horvitz, H. R. (2011). Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell, 147, 1525–1536.
Peters, A. H., O’Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schöfer, C., et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107, 323–337.
Petryk, N., Dalby, M., Wenger, A., Stromme, C. B., Strandsby, A., Andersson, R., & Groth, A. (2018). MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science (New York, N.Y.)., 361, 1389–1392.
Probst, A. V., Dunleavy, E., & Almouzni, G. (2009). Epigenetic inheritance during the cell cycle. Nature Reviews. Molecular Cell Biology., 10, 192–206.
Ranjan, R., Snedeker, J., & Chen, X. (2019). Asymmetric centromeres differentially coordinate with mitotic machinery to ensure biased sister chromatid segregation in germline stem cells. Cell Stem Cell, 25(666–681), e665.
Reverón-Gómez, N., González-Aguilera, C., Stewart-Morgan, K. R., Petryk, N., Flury, V., Graziano, S., et al. (2018). Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Molecular Cell., 72, 239-249.e235.
Scharf, A. N., Meier, K., Seitz, V., Kremmer, E., Brehm, A., & Imhof, A. (2009). Monomethylation of lysine 20 on histone H4 facilitates chromatin maturation. Molecular and Cellular Biology, 29, 57–67.
Schlissel, G., & Rine, J. (2019). The nucleosome core particle remembers its position through DNA replication and RNA transcription. Proceedings of the National Academy Sciences., 116, 20605–20611.
Serra-Cardona, A., & Zhang, Z. (2018). Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends in Biochemical Sciences, 43, 136–148.
Stewart-Morgan, K. R., Petryk, N., & Groth, A. (2020). Chromatin replication and epigenetic cell memory. Nature Cell Biology, 22, 361–371.
Stillman, B. (2018). Histone modifications: insights into their influence on gene expression. Cell, 175, 6–9.
Tran, V., Feng, L., & Chen, X. (2013). Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions. Chromosome Research., 21, 255–269.
Tran, V., Lim, C., Xie, J., & Chen, X. (2012). Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science (New York, N.Y.)., 338, 679–682.
Wooten, M., Ranjan, R., & Chen, X. (2020). Asymmetric histone inheritance in asymmetrically dividing stem cells. Trends in Genetics, 36, 30–43.
Wooten, M., Snedeker, J., Nizami, Z. F., Yang, X., Ranjan, R., Urban, E., et al. (2019). Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nature Structural & Molecular Biology., 26, 732–743.
Xie, J., Wooten, M., Tran, V., Chen, B.-C., Pozmanter, C., Simbolon, C., et al. (2015). Histone H3 threonine phosphorylation regulates asymmetric histone inheritance in the drosophila male germline. Cell, 163, 920–933.
Xie, J., Wooten, M., Tran, V., & Chen, X. (2017). Breaking symmetry–asymmetric histone inheritance in stem cells. Trends in Cell Biology., 27, 527–540.
Xu, M., Long, C., Chen, X., Huang, C., Chen, S., & Zhu, B. (2010). Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science (New York, N.Y.)., 328, 94–98.
Yu, C., Gan, H., Serra-Cardona, A., Zhang, L., Gan, S., Sharma, S., et al. (2018). A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science (New York, N.Y.)., 361, 1386–1389.
Yu, C., Gan, H., Han, J., Zhou, Z.-X., Jia, S., Chabes, A., et al. (2014). Strand-Specific Analysis Shows Protein Binding at Replication Forks and PCNA Unloading from Lagging Strands when Forks Stall. Molecular Cell, 56(4), 551–563.
Zhang, H., Gan, H., Wang, Z., Lee, J. H., Zhou, H., Ordog, T., et al. (2017). RPA interacts with HIRA and regulates H3.3 deposition at gene regulatory elements in mammalian cells. Molecular Cell, 65, 272–284.
Author information
Qing Wen and Yuan Yao have contributed equally to this work.
Affiliations
Shenzhen Key Laboratory of Synthetic Genomics, Guangdong Provincial Key Laboratory of Synthetic Genomics, CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, 518055, China
Qing Wen, Yuan Yao, Xinran Li, Zheng Hu, Hui Mei & Haiyun Gan
Corresponding author
Correspondence to Haiyun Gan.
Rights and permissions
About this article
Cite this article
Wen, Q., Yao, Y., Li, X. et al. Chromatin replication and parental histone allocation. GENOME INSTAB. DIS. 2, 51–58 (2021). https://doi.org/10.1007/s42764-021-00033-2
Received30 December 2020
Revised13 January 2021
Accepted22 January 2021
Published04 February 2021
Issue DateFebruary 2021
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable linkKeywords
DNA replication
Histone allocation
Nucleosome assembly
Epigenetic inheritance
Cell fate








用户登录
还没有账号?
立即注册