Fanconi anemia pathway and its relationship with cancer
Review Article
Open Access
Chenchen Dan, Hongjing Pei, Buzhe Zhang, Xuan Zheng, Dongmei Ran & Changzheng Du
Genome Instability & Disease 2, 175–183 (2021)
Abstract
Fanconi Anemia (FA) is a rare inherited hematological disease, caused by mutations in genes involved in the DNA interstrand crosslink (ICL) repair. Up to date, 22 genes have been identified that encode a series of functionally associated proteins that recognize ICL lesion and mediate the activation of the downstream DNA repair pathway including nucleotide excision repair, translesion synthesis, and homologous recombination. The FA pathway is strictly regulated by complex mechanisms such as ubiquitination, phosphorylation, and degradation signals that are essential for the maintenance of genome stability. Here, we summarize the discovery history and recent advances of the FA genes, and further discuss the role of FA pathway in carcinogenesis and cancer therapies.
Introduction
Fanconi anemia (FA) is a rare hereditary disease featured by a series of clinical manifestations including bone marrow failure, microcephaly, and absence or hypoplasia of radius and/or thumb (Auerbach, 2009); and the cells from FA patients exhibit characteristic hypersensitivity to DNA cross-link agents (Schneider et al., 2015). The clinical syndrome is widely distributed across different countries and populations, and is estimated to be 1 in every 100,000 newborns in the West (Nalepa & Clapp, 2018).
Fanconi anemia was named after the Swiss pediatrician Guido Fanconi who firstly described a case of this syndrome in 1927 (Lobitz & Velleuer, 2006). During his training as a pediatric at the children’s hospital of the University of Zurich, Guido Fanconi encountered a family in which three brothers died of a disease resembling pernicious anemia with a variety of congenital abnormalities but without signs of typically increased hemolysis. Following several other case reports, speculations about the etiology and hereditary pattern of this disease rises. Nearly 40 years after Fanconi’s description of the disorder, the enthusiasm of exploring the chromosomal abnormalities drove researchers to discover that the cells of FA patients had an elevated rate of chromosomal aberrations (Swift & Hirschhorn, 1966), and the FA was proved to be an autosomal recessive hereditary disorder (except one subtype which is X-linked) (Nalepa & Clapp, 2018).
The first identified FA gene is FANCC which accounts for about 14% of Fanconi anemia cases (Dong et al., 2015) and its c.711 + 4A > T is a dominant mutation in Ashkenazi Jews patients (Kutler & Auerbach, 2004). FANCC gene was successfully identified and its cDNA was cloned nearly 65 years after Guido Fanconi’s first case report (Strathdee et al., 1992). Then after 4 years since the first FANCC gene was cloned, the FANCA gene was cloned and reported, which was the second identified FA gene (Lo Ten Foe et al., 1996). Mutations in FANCA is responsible for the largest percentage of Fanconi anemia cases (over 65%), more than 500 pathogenic variants of FANCA gene have been determined in the Fanconi Anemia Mutation Database (Dong et al., 2015). FANCE and FANCF were identified with complementation cloning method in 2000 (b; de Winter, Leveille, et al., 2000). They are together responsible for 8% of mutations in Fanconi anemia cases (Schneider et al., 2015). FANCG gene was found to be identical with previously discovered human XRCC9 (X-ray repair, complementing defective repair in Chinese hamster cells 9), which was derived from the MMC-sensitive Chinese hamster mutant UV40 and was thought to be functional in the process of DNA post-replication repair (Winter et al., 1998). The other identified FA genes account for a small fraction (about 5%) of Fanconi anemia cases (Dong et al., 2015). As the knowledge to FA pathway develops, more and more genes were included into the FA family, such as BRCA1/2, PALB2, RAD51 and others (Table 1). Up to now, 22 FA genes have been identified, as listed in Table 1; more FA genes would be identified in the future since the known mutations in the FA family still could not shed light on all the FA cases till now.
Table 1 Fanconi anemia genes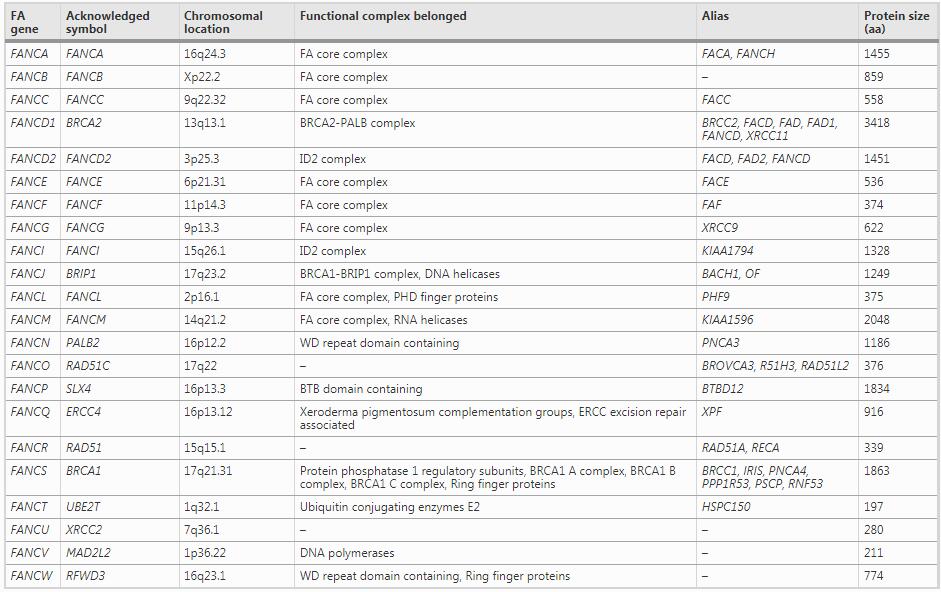
The function and working mechanism of FA pathway
FA pathway involves a bunch of FA proteins and other associated proteins, constituting a system serving for inter-strand cross-link (ICL) DNA repair in the S phase (Ceccaldi et al., 2016; Milletti et al., 2020), which coordinates each critical step by interacting with various DNA damage repair (DDR) proteins (Kottemann & Smogorzewska, 2013). ICL is a special type of DNA lesions that are a covalent linkage between opposite strands of double-stranded DNA (Fig. 1A). This kind of lesion prevents separation of DNA helix, making replication and transcription disturbed (Deans & West, 2011). In addition, ICL is also responsible for chromosomal aberrations and lethal DNA breaks (Vogel et al., 1998). Generally, in G1 phase, ICL lesions could be recognized and repaired by nucleotide excision repair (NER) machinery (Sarkar et al., 2006); while in S phase, ICL causes the stalling of DNA replication forks, which leads to the activation of FA pathway and the initiation of the DDR system (McHugh et al., 2001; Noll et al., 2006). During the process of DNA replication, the replication fork stops at the ICL lesion, inducing the replicative helicase complexes (CMG) to detach from the DNA strands upon the effect of the BRCA1 (Long et al., 2014) (Fig. 1A). Encounter with ICL lesion might lead to the shutdown of the replication fork, and this process would trigger downstream response about DNA repair and activation of ATR, which is essential for the subsequent activation of FA proteins. The FA pathway starts with ATP-dependent DNA translocase FANCM, and histone-like MHF complex (MHF1-MHF2-FAAP24), which recognizes and binds to the DNA damage portion and recruits other FA proteins (Collis & Boulton, 2010; Ling et al., 2016; Xue et al., 2008) (Fig. 1A). In this process, FANCM is activated by ATR, and is recruited to the ICL portion depending on its DNA-binding partners FAAP24, BLM complex, and the translocase activity of itself (Ling et al., 2016; Meetei et al., 2003).
FANCM further recruits the FA core complex, which is composed of a series of FA proteins as shown in Fig. 1B and Table 1. FA core complex and FANCM-BLM complex coordinate to unwind the DNA and promote the repair of replication fork (Deans & West, 2009; Meetei et al., 2003). FA core complex then catalyzes the mono-ubiquitination of FANCI and FANCD2, two members of ID2 complex, which is the critical step to activate ICL repair (Fig. 1B). Although perfect ubiquitination requires an integrated FA core complex, this process only necessarily needs an L-B-100 complex (FANCL-FANCB-FAAP100) (Rajendra et al., 2014), within which, the E3 ubiquitin ligase FANCL coordinates with UBE2T (another ubiquitin E2 ligase), playing the major role for the ubiquitination reaction. Additionally, UBL-5 and PCNA-ub are both required for the monoubiquitylation of FANCD2 (Howlett et al., 2009). Notably, the ubiquitination process of ID2 complex is reversible, catalyzed by the deubiquitinase USP1 and UAF1, which are responsible for the decruitment of ID2 complex after repair (Nijman et al., 2005).
The ubiquitinated ID2 complex further recruits structure-specific endonuclease to excise the stalled replication fork, such as XPF and FAN1. XPF-ERCC1 is an efficient incision endonuclease whose preferred substrate is the splayed arms DNA structure caused by ICL (Zhang & Walter, 2014), and it could be recruited to ICL lesions by ubiquitinated FAND2 (Klein Douwel et al., 2014). Similar to XPF, FAN1 could also be recruited by ubiquitinated FANCD2 via its UBZ4-type ubiquitin binding domain (Kratz et al., 2010), but this is not compelling in some cases (Zhang & Walter, 2014). Another essential scaffold protein mediating the endonuclease recruitment to ICL lesions is SLX4 (FANCP). SLX4 works together with SLX4IP, a constitutive factor in the SLX4 complex (Zhang et al., 2019), to recruit multiple nucleases such as XPF-ERCC1, MUS81-EME1 andSLX1to the ICL lesions for incisions (Klein Douwel et al., 2014; Sridharan et al., 2003); within which, the XPF–ERCC1–SLX4-SLX1 (XESS) complex is indispensable for the endonuclease activity in ICL repair (Zhang & Walter, 2014; Zhang et al., 2019) (Fig. 1C). Although some former studies reported ubiquitinated FAND2 was responsible for the recruitment of SLX4 (Yamamoto et al., 2011), the detailed molecular mechanism remains controversial since some other evidence showed the recruitment of SLX4 to ICL was independent on FANCD2 (Lachaud et al., 2014).
After incision, the cross-linked DNA would hang on the other strand, then the translesion synthesis (TLS) process would be launched. In the TLS process, PCNA-ub and the FA core complex recruit TLS polymerases such as REV7 (FANCV), polymerase ζ and REV1 to form a polymerase complex (Ceccaldi et al., 2016; Waters et al., 2009). Among those polymerases, the REV1-Polζ complex plays a leading role, recruited by FAAP20 in the core complex (Kim et al., 2012). Compared to the other DNA replication polymerases, the TLS polymerases have a larger binding pocket to adapt the ICL adduct (Ceccaldi et al., 2016); moreover, they could recognize PCNA-ub by the ub-binding domain which was not possessed by the other replication polymerases (Burschowsky et al., 2011) (Fig. 1D). While TLS produces a nice double strand, the incised portion remains a double-strand break (DSB) waiting for homologous recombination (HR) pathway to repair it. To launch HR repair, FANCD2 cooperates with BRCA1 and MRE11 to recruit CtIP, a key resection factor (Andres & Williams, 2017; Ceccaldi et al., 2016), to direct MRN complex (MRE11, RAD50 and NBS1) to cut the DSB portion and produce a 3’ single-strand (ss) DNA tail to invade into the homologous DNA strands (Liu & Huang, 2016) (Fig. 1E). During the DNA recombination, the recombinase RAD51 is responsible for forming the single-strand DNA nucleofilaments and strand invasion (Prakash et al., 2015) (Fig. 1F), which is recruited by BRCA2 (FANCD1)–PALB2 complex (Long et al., 2011; Shahid et al., 2014). Then a nascent DNA strand is synthesized by HR pathway to form an intact DNA duplex (Fig. 1F).
Like homologous recombination repair pathway, the FA pathway is also cell cycle dependent. During the process, double-strand break is accompanied by the ICL lesion excision, which is finally repaired by HR. Thus, a successful repair of ICL not only depends on normal FA genes, but also a intact HR pathway.
Association of FA genes with cancer
In clinic, a major cause for the death of FA patients is the cancer development in addition to bone marrow failure. A large population survey including 111 FA patients indicates a cancer frequency of 30%, with a median onset age before 20 (Steinberg-Shemer et al., 2020). Myeloid leukemias, liver tumors, head and neck carcinomas, and gynecologic malignancies are the most profound predisposing cancers among FA patients (Niraj et al., 2019). In clinical tumor tissues, either familial or sporadic cancer, the FA gene mutations are also common (Table 2), suggesting a intrinsic relationship between FA pahway and carcinogenesis. The reason behind is the critical function of FA pathway in maintaining genome stability. Deficient FA pathway causes genome instability resulting in accumulation of mutations, which gives additional survival advantages for the FA gene mutated cells, accerlerating the carcinogenesis process for some kinds of malignant tumors. For example, deletion and low expression of FANCA is common in acute myelocytic leukemia (AML), prostate cancer and oral cancer (Mantere et al., 2015; Tischkowitz et al., 2004; Tremblay et al., 2006). Similarly, heterozygous mutation of FANCD2 was confirmed to be associated with esophageal cancer, childhood T-cell acute lymphoblastic leukemia (ALL) and testicular seminoma (Akbari et al., 2011; Smetsers et al., 2012). Another well-known example is BRCA1 and BRCA2 (FANCS and FANCD1), whose germline mutations lead to familial breast and ovarian cancer (Antoniou et al., 2003; Chen & Parmigiani, 2007). The breast cancer appears more relevant to FA pathway: except BRCA1/2, germline mutation of RAD51C (FANCO) is also associated with an increased risk of breast cancer (Levy-Lahad, 2010; Loveday et al., 2012), and FANCM mutation increases the susceptibility of triple-negative breast cancer (Kiiski et al., 2014). The relationship between FA gene mutations and cancer predisposition is summarized in Table 2.
Table 2 FA gene mutations and cancer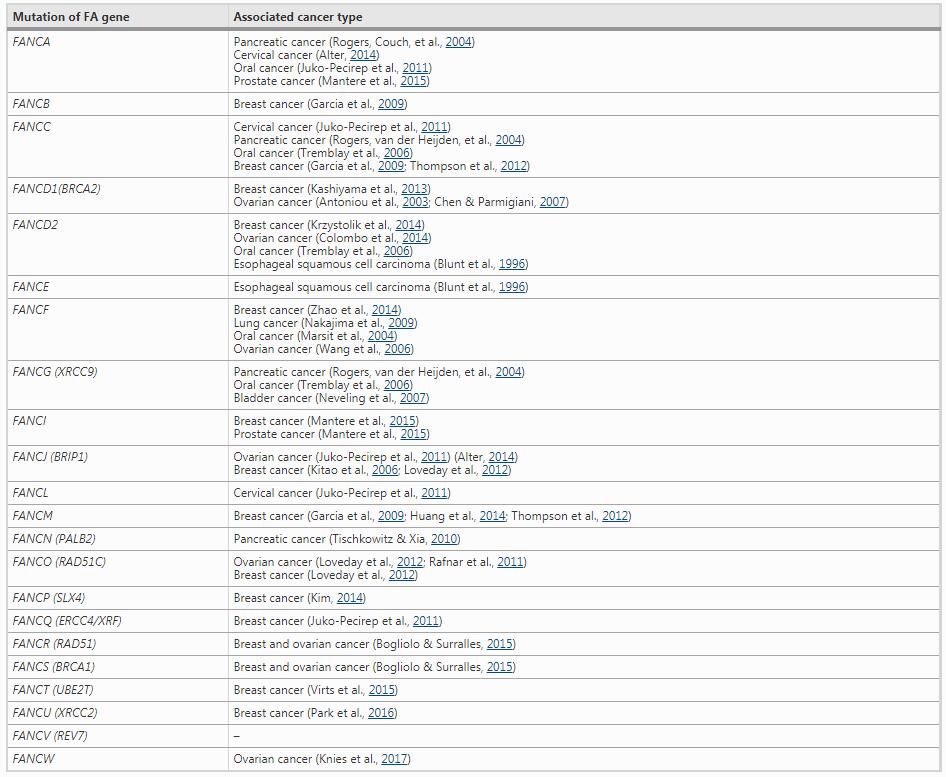
The epigenetic aberrance such as DNA methylation of FA genes is also common in cancers, and some of them have cancer-type specificity. For example, the hypermethylation of CpG islands in the promoter regions of FANCC and FANCL were found in AML and ALL (Hess et al., 2008), whereas the other FA genes were rarely methylated in the leukemia (Meyer et al., 2006). In solid tumors, hypermethylation of FANCF was found in multiple cancer types. In sporadic lung adenocarcinoma, FANCF hypermethylation is a predictor of poor survival compared to those with hypomethlylation (Marsit et al., 2004; Nakajima et al., 2009). In cervical cancer (CC), hypermethylation of BRCA1 and FANCF co-excist in patient samples, suggesting an attenuating function of FA pathway in CC (Narayan et al., 2004). In sporadic breast cancer, low expression of FANCD2 is commonly observed, which is associated with an improved long-term therapeutic outcome (Feng & Jin, 2019).
With regard to cancer therapy, the FA pathway is a ideal target. Early studies demonstrated that elevation of FA gene expression is frequently associated with chemo-resistance, whereas disruption of FA pathway sentitizes tumor cells to ICL agents (Chen et al., 2007; Taniguchi et al., 2003). For example, enhanced FA pathway activation has been shown to be relevant to resistance to melphalan in multiple myeloma and pancreatic cancers (Liu et al., 2020). An A549 derived cisplatin resistant lung cancer cell line A549/DR exhibits overexpression of multiple FA genes and elevated FANCD2 monoubiquitination level compared to its parental cell line (Liu et al., 2020). Knocking down of FA genes re-sensitizes A549/DR cells to cisplatin treatments. Similarly, another study revealed that knocking down of FANCF attenuates invasiveness of breast cancer, and improves the cellular sensitivity to the DSB agents doxorubicin (Zhao et al., 2014).
Since FA genes exert their critical function in maintaining genome stability by DNA damage repair, researchers have developed numerous therapies targeting the FA-deficient tumors. For example, a study revealed that the FA-deficient tumors present higher sensitivity to ATM inhibitors (Kennedy et al., 2007), since these tumors depend on the compensatory roles of ATM on DNA damage repair. Another successful example is the PARP inhibitor, which is highly effective in BRCA1 or BRCA2-deficient tumors based on their defect homologous recombination repair function (Bryant et al., 2005; Nalepa & Clapp, 2018). Since both chemotherapy re-sensitization and synthetic lethality will benefit from FA pathway inhibition, multiple approaches have been employed for the development of FA-specific inhibitors, but only a few of FA proteins including FANCL, FANCM, FANCR and FANCV have successful specific inhibitors (Brouwer et al., 2018; Hara et al., 2017; Lu et al., 2019). Some of the inhibitors exhibit excellent anti-cancer effect in preclinical study (Liu et al., 2020), while more FA inhibitors are still in demand. On the other hand, inhibiting FA proteins in cancer therapy is a double-edge sword. As the FA pathway mediates tumor suppression, targeting the FA pathway may further enhance genomic instability in tumor, which leads to increased genetic diversity and intratumoral heterogeneity, potentially promoting tumor progression and drug resistance after long-term treatment (Andor et al., 2017; Dagogo-Jack & Shaw, 2018). Another concern of FA inhibitors is it may bring other health issues, including anemia and a secondary tumor after long-term use, since genomic instability may promote carcinogenesis in healthy organs or tissues. Thus the clinical use of FA inhibitors needs cautious verification. Above all, although targeting FA pathway is a promising in cancer therapy, it remains a long way to transfer the lab findings into clinical practice.
Conclusion
As the rapid development of molecular biological research, people are increasingly understanding FA genes and their function. It is well established that the FA pathway plays a central role in ICL repair during which the FA proteins coordinate other DDR proteins to ensure the repair process and the genome stability. In the absence of FA genes, cells are predisposed to spontaneous DNA damage, leading to additional chromosomal aberrations which is closely associated with pernicious anemia and cancer. Understanding the molecular mechanism of FA pathway and other DNA repair pathways is greatly helpful for advancing cancer research. Moreover, novel treatment targeting FA pathway may play an important role for the development of personalized therapies to improve the clinical outcome of cancer patients.
Fig. 1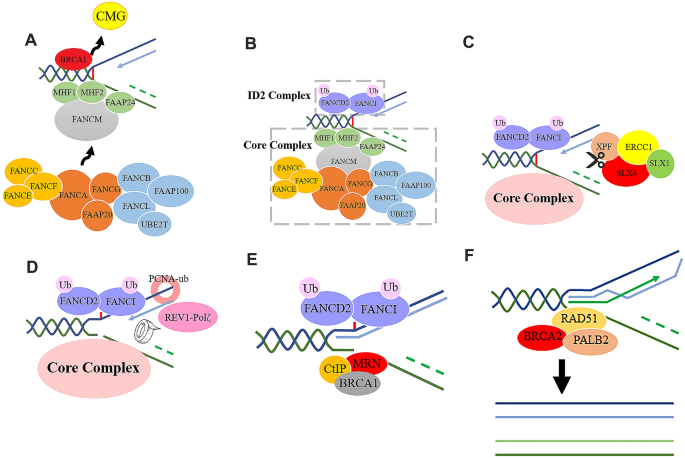
A As the replication fork converging with the ICL lesion, the CMG complex unfixes from DNA strands. The FANCM and MHF complex recognize the ICL lesion, binding with the stalled forks and recruiting the FA core complex; B the FA core complex recruits ID2 complex, and catalyze the mono-ubiquitination of FANCD2 and FANCI; C a series of structure-specific endonucleases are recruited to the ICL lesion for incision; D after incision, the TLS-polymerases such as REV1-Polζ are recruited by the FA core complex and PCNA-ub to initiate the TLS repair process; E since the TLS has provided an intact DNA double-helix as a template, MRN complex bind to the DSB portion of the other DNA strand (deep green) under the mediation of BRCA1 and CtIP, to initiate the HR repair pathway based on the nascent DNA template; F the HR process mediated by RAD51 and other DDR proteins synthesizes a nascent DNA strand using the TLS provided DNA double strands as a template. ICL interstrand crosslink, TLS translesion synthesis, DSB double strand break, HR homologous recombination, DDR DNA damage repair
Full size imageReferences
Akbari, M. R., Malekzadeh, R., Lepage, P., Roquis, D., Sadjadi, A. R., Aghcheli, K., Yazdanbod, A., Shakeri, R., Bashiri, J., Sotoudeh, M., et al. (2011). Mutations in Fanconi anemia genes and the risk of esophageal cancer. Human Genetics, 129(5), 573–582.
Alter, B. P. (2014). Fanconi anemia and the development of leukemia. Best Practice and Research Clinical Haematology, 27(3–4), 214–221.
Andor, N., Maley, C. C., & Ji, H. P. (2017). Genomic instability in cancer: Teetering on the limit of tolerance. Cancer Research, 77(9), 2179–2185.
Andres, S. N., & Williams, R. S. (2017). CtIP/Ctp1/Sae2, molecular form fit for function. DNA Repair (amst), 56, 109–117.
Antoniou, A., Pharoah, P. D. P., Narod, S., Risch, H. A., Eyfjord, J. E., Hopper, J. L., Loman, N., Olsson, H., Johannsson, O., Borg, A., et al. (2003). Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: A combined analysis of 22 studies. American Journal of Human Genetics, 72(5), 1117–1130.
Auerbach, A. D. (2009). Fanconi anemia and its diagnosis. Mutation Research, 668(1–2), 4–10.
Blunt, T., Gell, D., Fox, M., Taccioli, G. E., Lehmann, A. R., Jackson, S. P., & Jeggo, P. A. (1996). Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proceedings of the National Academy of Sciences of the United States of America, 93(19), 10285–10290.
Bogliolo, M., & Surralles, J. (2015). Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Current Opinion in Genetics and Development, 33, 32–40.
Brouwer, I., Moschetti, T., Candelli, A., Garcin, E. B., Modesti, M., Pellegrini, L., Wuite, G. J., & Peterman, E. J. (2018). Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA. The EMBO Journal, 37(7), 98162.
Bryant, H. E., Schultz, N., Thomas, H. D., Parker, K. M., Flower, D., Lopez, E., Kyle, S., Meuth, M., Curtin, N. J., & Helleday, T. (2005). Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature, 434(7035), 913–917.
Burschowsky, D., Rudolf, F., Rabut, G., Herrmann, T., Peter, M., & Wider, G. (2011). Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin. Journal of Biological Chemistry, 286(2), 1364–1373.
Ceccaldi, R., Sarangi, P., & D’Andrea, A. D. (2016). The Fanconi anaemia pathway: new players and new functions. Nature Reviews Molecular Cell Biology, 17(6), 337–349.
Chen, C. C., Taniguchi, T., & D’Andrea, A. (2007). The Fanconi anemia (FA) pathway confers glioma resistance to DNA alkylating agents. Journal of Molecular Medicine, 85(5), 497–509.
Chen, S., & Parmigiani, G. (2007). Meta-analysis of BRCA1 and BRCA2 penetrance. Journal of Clinical Oncology, 25(11), 1329–1333.
Collis, S. J., & Boulton, S. J. (2010). FANCM: fork pause, rewind and play. EMBO Journal, 29(4), 703–705.
Colombo, P. E., Fabbro, M., Theillet, C., Bibeau, F., Rouanet, P., & Ray-Coquard, I. (2014). Sensitivity and resistance to treatment in the primary management of epithelial ovarian cancer. Critical Reviews in Oncology Hematology, 89(2), 207–216.
Dagogo-Jack, I., & Shaw, A. T. (2018). Tumour heterogeneity and resistance to cancer therapies. Nature Reviews Clinical Oncology, 15(2), 81–94.
de Winter, J. P., Leveille, F., van Berkel, C. G., Rooimans, M. A., van Der Weel, L., Steltenpool, J., Demuth, I., Morgan, N. V., Alon, N., Bosnoyan-Collins, L., et al. (2000). Isolation of a cDNA representing the Fanconi anemia complementation group E gene. American Journal of Human Genetics, 67(5), 1306–1308.
de Winter, J. P., Rooimans, M. A., van Der Weel, L., van Berkel, C. G., Alon, N., Bosnoyan-Collins, L., de Groot, J., Zhi, Y., Waisfisz, Q., Pronk, J. C., et al. (2000). The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nature Genetics, 24(1), 15–16.
de Winter, J. P., Waisfisz, Q., Rooimans, M. A., van Berkel, C. G., Bosnoyan-Collins, L., Alon, N., Carreau, M., Bender, O., Demuth, I., Schindler, D., et al. (1998). The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nature Genetics, 20(3), 281–283.
Deans, A. J., & West, S. C. (2009). FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Molecular Cell, 36(6), 943–953.
Deans, A. J., & West, S. C. (2011). DNA interstrand crosslink repair and cancer. Nature Reviews Cancer, 11(7), 467–480.
Dong, H., Nebert, D. W., Bruford, E. A., Thompson, D. C., Joenje, H., & Vasiliou, V. (2015). Update of the human and mouse Fanconi anemia genes. Human Genomics, 9, 32.
Feng, L., & Jin, F. (2019). Expression and prognostic significance of Fanconi anemia group D2 protein and breast cancer type 1 susceptibility protein in familial and sporadic breast cancer. Oncology Letters, 17(4), 3687–3700.
Garcia, M. J., Fernandez, V., Osorio, A., Barroso, A., Llort, G., Lazaro, C., Blanco, I., Caldes, T., de la Hoya, M., Ramon, Y. C. T., et al. (2009). Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Research and Treatment, 113(3), 545–551.
Hara, K., Taharazako, S., Ikeda, M., Fujita, H., Mikami, Y., Kikuchi, S., Hishiki, A., Yokoyama, H., Ishikawa, Y., Kanno, S. I., et al. (2017). Dynamic feature of mitotic arrest deficient 2-like protein 2 (MAD2L2) and structural basis for its interaction with chromosome alignment-maintaining phosphoprotein (CAMP). The Journal of Biological Chemistry, 292(43), 17658–17667.
Hess, C. J., Ameziane, N., Schuurhuis, G. J., Errami, A., Denkers, F., Kaspers, G. J., Cloos, J., Joenje, H., Reinhardt, D., Ossenkoppele, G. J., et al. (2008). Hypermethylation of the FANCC and FANCL promoter regions in sporadic acute leukaemia. Cellular Oncology, 30(4), 299–306.
Howlett, N. G., Harney, J. A., Rego, M. A., Kolling, F. W. T., & Glover, T. W. (2009). Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. Journal of Biological Chemistry, 284(42), 28935–28942.
Huang, F., Ben Aissa, M., Magron, A., Huard, C. C., Godin, C., Levesque, G., & Carreau, M. (2014). The Fanconi anemia group C protein interacts with uncoordinated 5A and delays apoptosis. PLoS ONE, 9(3), e92811.
Juko-Pecirep, I., Ivansson, E. L., & Gyllensten, U. B. (2011). Evaluation of Fanconi anaemia genes FANCA, FANCC and FANCL in cervical cancer susceptibility. Gynecologic Oncology, 122(2), 377–381.
Kashiyama, K., Nakazawa, Y., Pilz, D. T., Guo, C. W., Shimada, M., Sasaki, K., Fawcett, H., Wing, J. F., Lewin, S. O., Carr, L., et al. (2013). Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes cockayne syndrome, xeroderma pigmentosum, and fanconi anemia. American Journal of Human Genetics, 92(5), 807–819.
Kennedy, R. D., Chen, C. C., Stuckert, P., Archila, E. M., De la Vega, M. A., Moreau, L. A., Shimamura, A., & D’Andrea, A. D. (2007). Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. The Journal of Clinical Investigation, 117(5), 1440–1449.
Kiiski, J. I., Pelttari, L. M., Khan, S., Freysteinsdottir, E. S., Reynisdottir, I., Hart, S. N., Shimelis, H., Vilske, S., Kallioniemi, A., Schleutker, J., et al. (2014). Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proceeding of the National Academy of the Sciences of the United States of America, 111(42), 15172–15177.
Kim, H., Yang, K., Dejsuphong, D., & D’Andrea, A. D. (2012). Regulation of Rev1 by the Fanconi anemia core complex. Nature Structural & Molecular Biology, 19(2), 164–170.
Kim, Y. (2014). Nuclease delivery: versatile functions of SLX4/FANCP in genome maintenance. Molecules and Cells, 37(8), 569–574.
Kitao, H., Yamamoto, K., Matsushita, N., Ohzeki, M., Ishiai, M., & Takata, M. (2006). Functional interplay between BRCA2/FancD1 and FancC in DNA repair. Journal of Biological Chemistry, 281(30), 21312–21320.
Klein Douwel, D., Boonen, R. A., Long, D. T., Szypowska, A. A., Raschle, M., Walter, J. C., & Knipscheer, P. (2014). XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Molecular Cell, 54(3), 460–471.
Knies, K., Inano, S., Ramirez, M. J., Ishiai, M., Surralles, J., Takata, M., & Schindler, D. (2017). Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. The Journal of Clinical Investigation, 127(8), 3013–3027.
Kottemann, M. C., & Smogorzewska, A. (2013). Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature, 493(7432), 356–363.
Kratz, K., Schopf, B., Kaden, S., Sendoel, A., Eberhard, R., Lademann, C., Cannavo, E., Sartori, A. A., Hengartner, M. O., & Jiricny, J. (2010). Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell, 142(1), 77–88.
Krzystolik, K., Jakubowska, A., Gronwald, J., Krawczynski, M. R., Drobek-Slowik, M., Sagan, L., Cyrylowski, L., Lubinski, W., Lubinski, J., & Cybulski, C. (2014). Large deletion causing von Hippel-Lindau disease and hereditary breast cancer syndrome. Hered Cancer Clin Pract, 12(1), 16.
Kutler, D. I., & Auerbach, A. D. (2004). Fanconi anemia in Ashkenazi Jews. Familial Cancer, 3(3–4), 241–248.
Lachaud, C., Castor, D., Hain, K., Munoz, I., Wilson, J., MacArtney, T. J., Schindler, D., & Rouse, J. (2014). Distinct functional roles for the two SLX4 ubiquitin-binding UBZ domains mutated in Fanconi anemia. Journal of Cell Science, 127(Pt 13), 2811–2817.
Levy-Lahad, E. (2010). Fanconi anemia and breast cancer susceptibility meet again. Nature Genetics, 42(5), 368–369.
Ling, C., Huang, J., Yan, Z., Li, Y., Ohzeki, M., Ishiai, M., Xu, D., Takata, M., Seidman, M., & Wang, W. (2016). Bloom syndrome complex promotes FANCM recruitment to stalled replication forks and facilitates both repair and traverse of DNA interstrand crosslinks. Cell Discovery, 2, 16047.
Liu, T., & Huang, J. (2016). DNA End resection: Facts and mechanisms. Genomics, Proteomics & Bioinformatics, 14(3), 126–130.
Liu, W., Palovcak, A., Li, F., Zafar, A., Yuan, F., & Zhang, Y. (2020). Fanconi anemia pathway as a prospective target for cancer intervention. Cell & Bioscience, 10, 39.
Lo Ten Foe, J. R., Rooimans, M. A., Bosnoyan-Collins, L., Alon, N., Wijker, M., Parker, L., Lightfoot, J., Carreau, M., Callen, D. F., Savoia, A., et al. (1996). Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nature Genetics, 14(3), 320–323.
Lobitz, S., & Velleuer, E. (2006). Guido Fanconi (1892–1979): a jack of all trades. Nature Reviews Cancer, 6(11), 893–898.
Long, D. T., Joukov, V., Budzowska, M., & Walter, J. C. (2014). BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Molecular Cell, 56(1), 174–185.
Long, D. T., Raschle, M., Joukov, V., & Walter, J. C. (2011). Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science, 333(6038), 84–87.
Loveday, C., Turnbull, C., Ruark, E., Xicola, R. M., Ramsay, E., Hughes, D., Warren-Perry, M., Snape, K., Breast Cancer Susceptibility, C., Eccles, D., et al. (2012). Germline RAD51C mutations confer susceptibility to ovarian cancer. Nature Genetics, 44(5), 475–476.
Lu, R., O’Rourke, J. J., Sobinoff, A. P., Allen, J. A. M., Nelson, C. B., Tomlinson, C. G., Lee, M., Reddel, R. R., Deans, A. J., & Pickett, H. A. (2019). The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nature Communications, 10(1), 2252.
Mantere, T., Haanpaa, M., Hanenberg, H., Schleutker, J., Kallioniemi, A., Kahkonen, M., Parto, K., Avela, K., Aittomaki, K., von Koskull, H., et al. (2015). Finnish Fanconi anemia mutations and hereditary predisposition to breast and prostate cancer. Clinical Genetics, 88(1), 68–73.
Marsit, C. J., Liu, M., Nelson, H. H., Posner, M., Suzuki, M., & Kelsey, K. T. (2004). Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene, 23(4), 1000–1004.
McHugh, P. J., Spanswick, V. J., & Hartley, J. A. (2001). Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. The Lancet Oncology, 2(8), 483–490.
Meetei, A. R., Sechi, S., Wallisch, M., Yang, D., Young, M. K., Joenje, H., Hoatlin, M. E., & Wang, W. (2003). A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Molecular and Cellular Biology, 23(10), 3417–3426.
Meyer, S., White, D. J., Will, A. M., Eden, T., Sim, A., Brown, R., & Strathdee, G. (2006). No evidence of significant silencing of Fanconi genes FANCF and FANCB or Nijmegen breakage syndrome gene NBS1 by DNA hyper-methylation in sporadic childhood leukaemia. British Journal of Haematology, 134(1), 61–63.
Milletti, G., Strocchio, L., Pagliara, D., Girardi, K., Carta, R., Mastronuzzi, A., Locatelli, F., & Nazio, F. (2020). Canonical and noncanonical roles of fanconi anemia proteins: implications in cancer predisposition. Cancers, 12(9), 2684.
Nakajima, T., Yasufuku, K., Suzuki, M., Fujiwara, T., Shibuya, K., Takiguchi, Y., Hiroshima, K., Kimura, H., & Yoshino, I. (2009). Assessment of chemosensitivity-related aberrant methylation of nonsmall cell lung cancer by EBUS-TBNA. Journal of Bronchology Interventional Pulmonology, 16(1), 10–14.
Nalepa, G., & Clapp, D. W. (2018). Fanconi anaemia and cancer: an intricate relationship. Nature Reviews Cancer, 18(3), 168–185.
Narayan, G., Arias-Pulido, H., Nandula, S. V., Basso, K., Sugirtharaj, D. D., Vargas, H., Mansukhani, M., Villella, J., Meyer, L., Schneider, A., et al. (2004). Promoter hypermethylation of FANCF: disruption of Fanconi Anemia-BRCA pathway in cervical cancer. Cancer Research, 64(9), 2994–2997.
Neveling, K., Kalb, R., Florl, A. R., Herterich, S., Friedl, R., Hoehn, H., Hader, C., Hartmann, F. H., Nanda, I., Steinlein, C., et al. (2007). Disruption of the FA/BRCA pathway in bladder cancer. Cytogenetic and Genome Research, 118(2–4), 166–176.
Nijman, S. M., Huang, T. T., Dirac, A. M., Brummelkamp, T. R., Kerkhoven, R. M., D’Andrea, A. D., & Bernards, R. (2005). The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Molecular Cell, 17(3), 331–339.
Niraj, J., Farkkila, A., & D’Andrea, A. D. (2019). The Fanconi anemia pathway in cancer. Annual Review of Cancer Biology, 3, 457–478.
Noll, D. M., Mason, T. M., & Miller, P. S. (2006). Formation and repair of interstrand cross-links in DNA. Chemical Reviews, 106(2), 277–301.
Park, J. Y., Virts, E. L., Jankowska, A., Wiek, C., Othman, M., Chakraborty, S. C., Vance, G. H., Alkuraya, F. S., Hanenberg, H., & Andreassen, P. R. (2016). Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. Journal of Medical Genetics, 53(10), 672–680.
Prakash, R., Zhang, Y., Feng, W., & Jasin, M. (2015). Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harbor Perspectives in Biology, 7(4), 16600.
Rafnar, T., Gudbjartsson, D. F., Sulem, P., Jonasdottir, A., Sigurdsson, A., Jonasdottir, A., Besenbacher, S., Lundin, P., Stacey, S. N., Gudmundsson, J., et al. (2011). Mutations in BRIP1 confer high risk of ovarian cancer. Nature Genetics, 43(11), 1104–1107.
Rajendra, E., Oestergaard, V. H., Langevin, F., Wang, M., Dornan, G. L., Patel, K. J., & Passmore, L. A. (2014). The genetic and biochemical basis of FANCD2 monoubiquitination. Molecular Cell, 54(5), 858–869.
Rogers, C. D., Couch, F. J., Brune, K., Martin, S. T., Philips, J., Murphy, K. M., Petersen, G., Yeo, C. J., Hruban, R. H., & Goggins, M. (2004). Genetics of the FANCA gene in familial pancreatic cancer. Journal of Medical Genetics, 41(12), 126.
Rogers, C. D., van der Heijden, M. S., Brune, K., Yeo, C. J., Hruban, R. H., Kern, S. E., & Goggins, M. (2004). The genetics of FANCC and FANCG in familial pancreatic cancer. Cancer Biology & Therapy, 3(2), 167–169.
Sarkar, S., Davies, A. A., Ulrich, H. D., & McHugh, P. J. (2006). DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. The EMBO Journal, 25(6), 1285–1294.
Schneider, M., Chandler, K., Tischkowitz, M., & Meyer, S. (2015). Fanconi anaemia: genetics, molecular biology, and cancer—implications for clinical management in children and adults. Clinical Genetics, 88(1), 13–24.
Shahid, T., Soroka, J., Kong, E., Malivert, L., McIlwraith, M. J., Pape, T., West, S. C., & Zhang, X. (2014). Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nature Structural & Molecular Biology, 21(11), 962–968.
Smetsers, S., Muter, J., Bristow, C., Patel, L., Chandler, K., Bonney, D., Wynn, R. F., Whetton, A. D., Will, A. M., Rockx, D., et al. (2012). Heterozygote FANCD2 mutations associated with childhood T Cell ALL and testicular seminoma. Familial Cancer, 11(4), 661–665.
Sridharan, D., Brown, M., Lambert, W. C., McMahon, L. W., & Lambert, M. W. (2003). Nonerythroid alphaII spectrin is required for recruitment of FANCA and XPF to nuclear foci induced by DNA interstrand cross-links. Journal of Cell Science, 116(Pt 5), 823–835.
Steinberg-Shemer, O., Goldberg, T. A., Yacobovich, J., Levin, C., Koren, A., Revel-Vilk, S., Ben-Ami, T., Kuperman, A. A., Zemer, V. S., Toren, A., et al. (2020). Characterization and genotype-phenotype correlation of patients with Fanconi anemia in a multi-ethnic population. Haematologica, 105(7), 1825–1834.
Strathdee, C. A., Gavish, H., Shannon, W. R., & Buchwald, M. (1992). Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature, 356(6372), 763–767.
Swift, M. R., & Hirschhorn, K. (1966). Fanconi’s anemia. Inherited susceptibility to chromosome breakage in various tissues. Annals of Internal Medicine, 65(3), 496–503.
Taniguchi, T., Tischkowitz, M., Ameziane, N., Hodgson, S. V., Mathew, C. G., Joenje, H., Mok, S. C., & D’Andrea, A. D. (2003). Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nature Medicine, 9(5), 568–574.
Thompson, E. R., Doyle, M. A., Ryland, G. L., Rowley, S. M., Choong, D. Y., Tothill, R. W., Thorne, H., Barnes, D. R., Li, J., et al. (2012). Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genetics, 8(9), e1002894.
Tischkowitz, M. D., Morgan, N. V., Grimwade, D., Eddy, C., Ball, S., Vorechovsky, I., Langabeer, S., Stoger, R., Hodgson, S. V., & Mathew, C. G. (2004). Deletion and reduced expression of the Fanconi anemia FANCA gene in sporadic acute myeloid leukemia. Leukemia, 18(3), 420–425.
Tischkowitz, M., & Xia, B. (2010). PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Research, 70(19), 7353–7359.
Tremblay, S., dos Reis, P. P., Bradley, G., Galloni, N. N., Perez-Ordonez, B., Freeman, J., Brown, D., Gilbert, R., Gullane, P., Irish, J., et al. (2006). Young patients with oral squamous cell carcinoma—study of the involvement of GSTP1 and deregulation of the Fanconi anemia genes. Archives of Otolaryngology, 132(9), 958–966.
Virts, E. L., Jankowska, A., Mackay, C., Glaas, M. F., Wiek, C., Kelich, S. L., Lottmann, N., Kennedy, F. M., Marchal, C., Lehnert, E., et al. (2015). AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Human Molecular Genetics, 24(18), 5093–5108.
Vogel, E. W., Barbin, A., Nivard, M. J., Stack, H. F., Waters, M. D., & Lohman, P. H. (1998). Heritable and cancer risks of exposures to anticancer drugs: inter-species comparisons of covalent deoxyribonucleic acid-binding agents. Mutation Research, 400(1–2), 509–540.
Wang, Z., Li, M., Lu, S., Zhang, Y., & Wang, H. (2006). Promoter hypermethylation of FANCF plays an important role in the occurrence of ovarian cancer through disrupting Fanconi anemia-BRCA pathway. Cancer Biology & Therapy, 5(3), 256–260.
Waters, L. S., Minesinger, B. K., Wiltrout, M. E., D’Souza, S., Woodruff, R. V., & Walker, G. C. (2009). Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiology and Molecular Biology Reviews, 73(1), 134–154.
Xue, Y., Li, Y., Guo, R., Ling, C., & Wang, W. (2008). FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Human Molecular Genetics, 17(11), 1641–1652.
Yamamoto, K. N., Kobayashi, S., Tsuda, M., Kurumizaka, H., Takata, M., Kono, K., Jiricny, J., Takeda, S., & Hirota, K. (2011). Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proceedings of the National Academy of Sciences of the United States of America, 108(16), 6492–6496.
Zhang, H., Chen, Z., Ye, Y., Ye, Z., Cao, D., Xiong, Y., Srivastava, M., Feng, X., Tang, M., Wang, C., et al. (2019). SLX4IP acts with SLX4 and XPF-ERCC1 to promote interstrand crosslink repair. Nucleic Acids Research, 47(19), 10181–10201.
Zhang, J., & Walter, J. C. (2014). Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair, 19, 135–142.
Zhao, L., Li, N., Yu, J. K., Tang, H. T., Li, Y. L., He, M., Yu, Z. J., Bai, X. F., Zheng, Z. H., Wang, E. H., et al. (2014). RNAi-mediated knockdown of FANCF suppresses cell proliferation, migration, invasion, and drug resistance potential of breast cancer cells. Brazilian Journal of Medical and Biological Research, 47(1), 24–34.
Acknowledgements
Chenchen Dan, Hongjing Pei and Buzhe Zhang do equal contribution for this work.
Author information
Affiliations
School of Medicine, The Southern University of Science and Technology, 1088 Xueyuan Avenue, Shenzhen, 518055, Guangdong, People’s Republic of China
Chenchen Dan, Hongjing Pei, Buzhe Zhang, Xuan Zheng, Dongmei Ran & Changzheng Du
Pathology department, Southern University of Science and Technology Hospital, Shenzhen, Guangdong, People’s Republic of China
Dongmei Ran
General Surgery Department, Southern University of Science and Technology Hospital, Shenzhen, Guangdong, People’s Republic of China
Changzheng Du
Corresponding author
Correspondence to Changzheng Du.
Ethics declarations
Conflict of interest
There is no conflict of interests among the authors.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dan, C., Pei, H., Zhang, B. et al. Fanconi anemia pathway and its relationship with cancer. GENOME INSTAB. DIS. 2, 175–183 (2021). https://doi.org/10.1007/s42764-021-00043-0
Received07 April 2021
Accepted22 May 2021
Published05 June 2021
Issue DateJune 2021
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable linkKeywords
Fanconi anemia
Genes
Cancer
Human








用户登录
还没有账号?
立即注册