SIRT7 is a deacetylase of N4-acetylcytidine on ribosomal RNA
Short Communication
Open Access
Genome Instability & Disease 2, 253–260 (2021)
Abstract
N-acetyltransferase 10 catalyzes RNA N4-acetylcytidine (ac4C) modifications and thus regulates RNA stability and translation efficiency. However, the deacetylase for ac4C is unknown. SIRT7 was initially identified as an NAD+-dependent protein deacetylase and plays essential roles in genome stability, circadian rhythms, metabolism, and aging. In this study, we identified SIRT7 as a deacetylase of the ac4C of ribosomal (r)RNA for the first time and found it to be NAD+-independent. Our data highlight the important role of SIRT7 in rRNA ac4C modification and suggest an additional epitranscriptional regulation of aging.
Introduction
RNAs can be covalently modified, e.g., N1-methyladenosine, N4-acetylcytidine (ac4C), N6-methyladenosine, N7-methylguanosine, and 5-methylcytidine, etc., most of which regulate the stability and translation efficiency of the target RNA (Wiener & Schwartz, 2021). Ac4C modification was first identified in yeast transfer (t)RNA almost 5 decades ago (Kowalski et al., 1971; Zachau et al., 1966). In 2014, Tsutomu Suzuki’s team found that KRE33, the Saccharomyces cerevisiae homolog of human N-acetyltransferase 10 (NAT10), catalyzes the ac4C1773 of 18S ribosomal (r)RNA, promoting the formation of pre-18S rRNA (Sharma et al., 2015). In humans, NAT10 catalyzes the ac4C1842 of 18S rRNA (Ito et al., 2014a, 2014b). Using an antibody that recognizes ac4C, Oberdoerffer’s group expanded the scope of ac4C modification targets to mRNA, the upper part of which is extensively ac4C-modified by NAT10 thus to regulate mRNA stability and translation efficiency (Arango et al., 2018; Sinclair et al., 2017). Like protein post-translational modifiers, the RNA-modifying enzymes and related factors are also categorized to three groups: writers, erasers, and readers. The identity of the ac4C eraser is undetermined (Jin et al., 2020).
Sirtuins are NAD+-dependent deacylases and mono-ADP-ribosyltransferases that target histones and numerous nonhistone substrates (O'Callaghan & Vassilopoulos, 2017). Of the seven mammalian sirtuins, SIRT7 is the only one that localizes in the nucleolus. So far, dozens of protein deacetylation targets of SIRT7, including NPM1 (Ianni et al., 2021; Lee et al., 2014), CRY1 (Liu et al., 2019), GABP-β1 (Ryu et al., 2014), SMAD4 (Tang et al., 2017b), NFATC1 (Li et al., 2020), USP39 (Dong et al., 2020), PCAF (Lu et al., 2020), and H3K36 (Wang et al., 2019) etc., have been identified, and dictate the pleiotropic function of SIRT7 in genomic stability, lipid metabolism, circadian rhythms, hair regeneration, and aging (Tang, 2015; Yoshizawa et al., 2014). Interestingly, SIRT7 binds to nascent pre-rRNA, deacetylates PAF53, and enhances the rDNA occupancy of RNA polymerase I (Chen et al., 2013; Ford et al., 2006). SIRT7 interacts with mTOR and GTF3C1, and thus, regulates tRNA transcription and protein synthesis (Tsai et al., 2014). The aim of this study was to determine if SIRT7 functions as a deacetylase for ac4C modification of rRNA. We tested the interaction of SIRT7 and NAT10, examined the effect of SIRT7 overexpression on RNA ac4C, and measured ac4C levels of 18S rRNA in the presence or absence of SIRT7 and NAD+.
Materials and methods
Cell culture, plasmids and transfections
HEK293T and U2OS cells were cultured in DMEM medium (Corning) with 10% FBS (PAN) and 1% P/S (Gibco) at 37 °C in a humidified atmosphere under 5% CO2.
The following plasmids were used in this study: FLAG-SIRT1-7, FLAG-NAT10 and HA-SIRT7, which were constructed by cloning full-length cDNA into the pCDNA3.1 vector and indicated tags were added on the N-terminal. GFP-SIRT7 was constructed by cloning full-length SIRT7 cDNA into the pEGFP-C1 vector. GST-SIRT7, GST-SIRT7 (H187Y), GST-SIRT7 (RNAmut) and GST-SIRT7 (H187Y+RNAmut) were constructed by cloning full-length or mutant SIRT7 cDNA into the pGEX-4T-2 vector. The conserved catalytic base His187 of SIRT7 was substituted to Tyr in GST-SIRT7 (H187Y) mutant. The potential SIRT7 Protein-RNA binding motif 392KRTKRKK398 was mutated to 392AAAAAAA398 in GST-SIRT7 (RNAmut) mutant (Tong et al., 2017). GST-SIRT7 (H187Y+RNAmut) is double mutated with H187Y and RNAmut. Plasmid transfections were performed with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Recombinant protein preparation
The GST-SIRT7, -SIRT7 (H187Y), -SIRT7 (RNAmut), and -SIRT7 (H187Y+RNAmut) were expressed in E. coli BL21 (DE3) strain induced with IPTG (1 mM). Cell pellets were first resuspended in BC500 buffer (25 mM Tris–HCl pH7.3, 500 mM NaCl, 0.5% Triton X-100 and 20% glycerol) containing DTT (1 mM) and then were disrupted by sonication. Cleared cell lysates were incubated overnight with Glutathione Sepharose (GE Healthcare) at 4 ℃. The beads were washed with BC500 buffer once, and further washed with BC100 buffer (25 mM Tris–HCl pH7.3, 100 mM NaCl, 0.5% Triton X-100 and 20% glycerol) three times. Proteins were then eluted with Reduced Glutathione (Amresco) and stored at – 80 ℃.
Antibodies
The following antibodies were used in this study: Anti-FLAG M2(F3165, Sigma), Anti-NAT10 (ab194297, Abcam), Anti-HA Tag (3724, CST), Anti-Ack (PTM-105, PTM BIO), Anti-ac4C (ab252215, Abcam).
Immunoprecipitation and western blotting
HEK293T cells treated with the indicated transfections were harvested in lysis buffer [20 mM Tris–HCl (pH 7.5), 300 mM NaCl, 10% glycerol, 0.1 mM EDTA, 0.1% NP-40, and a complete protease inhibitor cocktail]. Cell lysates were incubated with 1 µg of the respective antibodies and then crosslinked to protein A/G agarose beads at 4 °C overnight with rotation. The bead-bound immunoprecipitates were washed with lysis buffer and boiled in SDS sample loading buffer. The inputs and immunoprecipitated products were resolved using SDS-PAGE and transferred to PVDF membranes (Millipore, USA). The membranes were blocked in 5% milk in Tris-buffered saline and Tween 20 [TBST: 150 mM NaCl, 20 mM Tris–HCl (pH 7.6), 0.05% Tween 20] for 1 h at room temperature and subjected to immunoblotting with the indicated antibodies overnight at 4 °C. The membranes were probed with Peroxidase-conjugated AffiniPure Goat Anti-Rabbit (or Mouse) IgG (H+L) (1:5000, Jackson ImmunoResearch Laboratories) for 1 h at room temperature, and visualized using an enhanced chemiluminescence kit (Pierce, USA) and in a Bio-Rad imaging system.
Immunofluorescence staining
U2OS cells transfected with the GFP-SIRT7 construct were fixed with 4% paraformaldehyde at 4 °C, then permeabilized with PBS containing 0.1% Triton X-100 for 15 min. Then, the cells were blocked with 1% bovine serum in PBS for 30 min at room temperature. The coverslips were incubated with anti-NAT10 antibody (1:200) overnight at 4 °C, followed by incubation with Alexa Fluor-conjugated secondary antibodies (1:500, Life Technologies) for 1 h at room temperature in the dark and mounted with DAPI-containing mounting medium. The images were captured under a confocal microscope (Zeiss, Germany).
In vitro transcription of ac4C-containing RNA probes
Complementary DNA (cDNA) was used as template for PCR amplification of 18S rRNA. 5′ primer contains the T7 promoter sequence 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′ (T7). To prepare templates for 18S rRNA, the following primer pair was used: 5′- (T7) TACCTGGTTGATCCTGCCAGTAGC-3′ and 5′-TAATGATCCTTCCGCAGGTTCACC-3′. ac4C-containing 18S rRNA probes were prepared using in vitro transcription of PCR-amplified DNA templates with T7 RNA polymerases (Thermo) as recommended. For the modified transcripts, cytidine triphosphate (CTP) was replaced with ac4CTP in the reaction mix. The reaction was incubated for 2–4 h at 37 °C, followed by incubation with DNaseI (Thermo; 1 U/1 µg of template DNA) for 30 min at 37 °C, and terminated by 10 mM EDTA with incubation at 65 °C for 10 min. Then the RNA probes were extracted with Trizol and stored at – 80 ℃. The incorporation of ac4C into the RNA probe was assessed by dot blot.
SIRT7 in vitro deacetylase assay
We preincubated 300 ng of ac4C-containing 18S rRNA probes with 2 μg of the purified GST-SIRT7, GST-SIRT7 (H187Y), GST-SIRT7 (RNAmut) and GST-SIRT7 (H187Y+RNAmut) proteins respectively in the following buffer: 10 mM Tris pH 8.0, 2 mM MgCl2, 0.2 mM DTT, and 10% glycerol either in the presence or absence of 2.5 mM NAD+. The reaction was carried out in a 30 ℃ water bath for 1 h. The RNA probe was then purified using Trizol and transferred onto Amersham Hybond-N+ membranes for further dot blot analysis.
ac4C detection by dot blotting
Dot blotting was performed using rabbit monoclonal anti-ac4C antibody (#ab252215, Abcam). Briefly, total RNA or in vitro transcribed probes were loaded onto Hybond-N+ membranes, crosslinked at 120,000 μJ/cm2, blocked with 5% non-fat milk in 0.1% Tween-20 TBS (TBST) for 30 min at room temperature, and probed overnight with the anti-ac4C antibody in 5% non-fat milk (1:1000) at 4 ℃. Membranes were washed four times with 0.1% TBST, incubated with Peroxidase-conjugated AffiniPure Goat Anti-Rabbit IgG (H+L) (1:5000, Jackson ImmunoResearch Laboratories) for 1 h at room temperature, and visualized using an enhanced chemiluminescence kit (Pierce, USA) on Bio-Rad imaging system.
ac4C detection by immuno-northern blotting
HEK293T cells were transfected with vector or FLAG-SIRT7 plasmids for 48 h and total RNAs were isolated. Equal amounts of total RNA (20 μg) were mixed with formaldehyde denaturing loading dye, incubated at 65℃ for 15 min and separated on 1% agarose denaturing gel containing nucleic acid dye. The gel was verified by UV imaging before transfer. RNA was transferred onto Hybond-N + membranes (GE Healthcare) by capillary transfer using 20×SSC buffer [3 M NaCl, 0.3 M sodium citrate, (pH 7.0)]. Membranes were then blotted with anti-ac4C antibodies.
18S rRNA degradation assay
To measure the degradation rate of 18S rRNA, HEK293T cells were transfected with vector, FLAG-SIRT7 plasmids, control or siSIRT7 siRNA for 48 h. The expression of 18S rRNA was shut off by adding actinomycin D (2 μg/ml) into the cell culture medium for 5 h, and total RNA was isolated and subjected to quantitative RT-PCR analysis. The primer sequences are as follows: 18S forward: 5′-CAGCCACCCGAGATTGAGCA-3′ and reverse: 5′-TAGTAGCGACGGGCGGTGTG-3′; GAPDH forward: 5′- CACCCACTCCTCCACCTTTG-3′ and reverse: 5′-CCACCACCCTGTTGCTGTAG-3′.
Statistical analysis
A two-tailed Student’s t test was used to determine the statistical significance of group differences. All data are presented as the means ± s.d. or means ± s.e.m. as indicated, and a p value of < 0.05 was considered statistically significant.
Results
SIRT7 interacts with NAT10
The ac4C acetylase activity of NAT10 is related to its acetylation state (Cai et al., 2017). We thus examined the interaction between sirtuins and NAT10 with the assumption that sirtuins might indirectly regulate ac4C level. As shown, western blotting of the anti-FLAG immunoprecipitates from HEK293T cells overexpressing FLAG-tagged sirtuins demonstrated that, of the seven sirtuin members, FLAG-SIRT7 had the highest NAT10-binding capacity (Fig. 1a). This was confirmed by detection of FLAG-NAT10 in anti-HA immunoprecipitates from HEK293T cells overexpressing FLAG-NAT10 with or without HA-SIRT7 (Fig. 1b). Reciprocally, HA-SIRT7 was observed in the anti-FLAG immunoprecipitates from HEK293T cells overexpressing HA-SIRT7 with or without FLAG-NAT10 (Fig. 1c). Further co-immunofluorescence staining indicated that NAT10 and GFP-SIRT7 co-localized in the nucleoli (Fig. 1d). We found that levels of pan-acetyl-lysine in the anti-NAT10 immunoprecipitates were comparable in the presence and absence of ectopic HA-SIRT7 (Fig. 1e), implicating that SIRT7 unlikely affects the ac4C level, if any, via deacetylating and modulating the acetylase activity of NAT10.
Fig. 1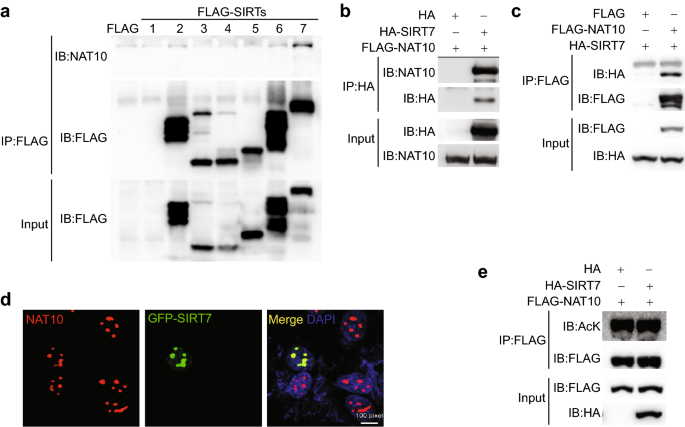
SIRT7 interacts and colocalizes with NAT10 in the nucleolus. a Representative immunoblots showing NAT10 in the anti-FLAG immunoprecipitates from HEK293T cells overexpressing FLAG-tagged sirtuins (SIRT1-7). b Representative immunoblots showing FLAG-NAT10 in the anti-HA immunoprecipitates from HEK293T cells overexpressing FLAG-NAT10 with or without HA-SIRT7. c Representative immunoblots showing HA-SIRT7 in the anti-HA immunoprecipitates from HEK293T cells overexpressing HA-SIRT7 and with or without FLAG-NAT10. d Representative immunofluorescence microscopic images showing colocalized endogenous NAT10 and ectopic GFP-SIRT7 in U2OS cells. e Representative immunoblots showing pan-acetyl-lysine (AcK) levels in the anti-FLAG-NAT10 immunoprecipitates from HEK293T cells overexpressing FLAG-NAT10 and with or without HA-SIRT7
Full size imageSIRT7 is a deacetylase of 18S rRNA ac4C
Next, we examined whether SIRT7 would directly affect the ac4C level of RNA. The overexpression of ectopic SIRT7 distinctly decreased the ac4C levels of the total RNA purified from HEK293T cells (Fig. 2a, b). To test whether SIRT7 is a deacetylase of ac4C, we purified recombinant human SIRT7, obtained 18S rRNA with incorporated ac4CTP via in vitro transcription, and carried out an in vitro deacetylation assay. The incubation with GST-SIRT7 significantly downregulated the levels of ac4C (Fig. 2c, d), and the presence of NAD+ only slightly reduced the ac4C level. The H187Y mutation of SIRT7 blocks its deacetylation activity towards protein targets (Mitra & Dey, 2020), and the SIRT7 mutation 392KRTKRKK398 (SIRT7 RNAmut) inhibits its binding to RNAs (Tong et al., 2017). Both SIRT7 H187Y and RNAmut significantly attenuated the deacetylase activity of ac4C on 18S rRNA, and an additive effect was observed with the double mutant (Fig. 2e, f). We next examined the effect of SIRT7 manipulation in mammalian cells. Overexpression of SIRT7 largely downregulated the ac4C level of 18S rRNA in HEK293T cells, determined by immuno-northern blotting (Fig. 2g). Further degradation assay revealed that SIRT7 overexpression significantly accelerated 18S rRNA degradation, while knockdown of SIRT7 significantly stabilized 18S rRNA (Fig. 2h). Together, these data support SIRT7 as a deacetylase of ac4C modification on 18S rRNA.
Fig. 2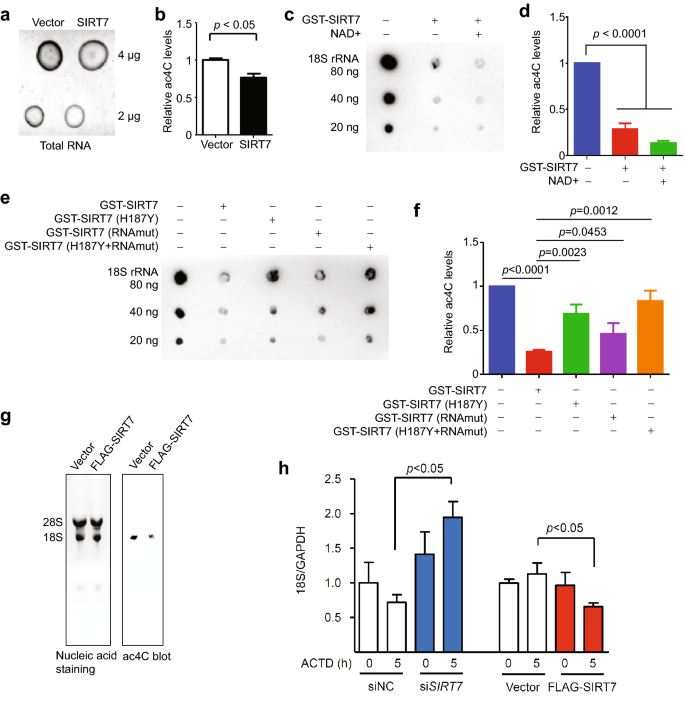
SIRT7 deacetylates ac4C rRNA. a Representative dot blot showing the levels of ac4C of total RNA from HEK293T cells with ectopic SIRT7 overexpression or mock (vector) treatment. b Quantification of a, showing significantly reduced ac4C levels in SIRT7-overexpressing cells. c Representative dot blot showing ac4C levels of 18S rRNA (in vitro transcribed to incorporate ac4CTP) in the presence or absence of GST-SIRT7 and/or NAD+ . d Quantification of c, showing significantly reduced ac4C levels in the presence of GST-SIRT7 regardless of NAD+. e Representative dot blot showing ac4C levels of 18S rRNA in the presence of indicated SIRT7 recombinant proteins. f Quantification of e, showing significantly reduced ac4C levels in the presence of GST-SIRT7. The effect was reduced in the case of the mutant SIRT7. g Representative images of agarose gel (left) and immune-northern blot (right) showing 18S rRNA level and ac4C level, respectively. h Quantitative RT-PCR analysis of 18S rRNA in HEK293T cells with indicated SIRT7 manipulations. ACTD, actinomycin D (2 μg/ml)
Full size imageDiscussion
Post-transcriptional modifications on RNAs, globally termed epitranscriptome, provide additional layers of response to environmental and endogenous stresses (Roundtree et al., 2017). Ac4C modification was evidenced decades ago, however, until recently and with the development of new molecular techniques, researchers have been able to reveal details of global ac4C; it has been mainly found in rRNA and tRNA, and with only a small fraction in mRNA (Sas-Chen et al., 2020; Thalalla Gamage et al., 2021). NAT10 is the only identified acetyltransferase that catalyzes ac4C modification. Given essential and common roles of SIRT7, as an acylation eraser, and of NAT10, as an ac4C RNA acetylation writer, in the regulation of aging, we speculated that SIRT7 may serve as an eraser of ac4C, and RNA ac4C might regulate aging. There was a clear decrease in the ac4C levels in the total RNA of HEK293T cells when SIRT7 was overexpressed. Both SIRT7 H187Y and RNAmut attenuated the deacetylase activity of ac4C on 18S rRNA. Thus, our data support the hypothesis that SIRT7 is a deacetylase of ac4C. We also found that the absence of NAD+ had little effect on the activity of SIRT7; therefore, its deacetylase activity appears to be NAD+-independent. Of note SIRT7 was previously reported to regulate the production of pre-rRNA and small nucleolar RNAs (snoRNAs) through the core components of the U3 snoRNP complex U3-55 k (Chen et al., 2016). We recently found that sirtuin SIRT6 recognizes double strand breaks and potentiates the DNA damage response (Meng et al., 2020). Whether SIRT6 and other sirtuins also deacetylate ac4C merits investigation.
Aging is accompanied by a gradual decline in sirtuin levels and/or activities, as well as NAD+ levels, making sirtuins some of the most viable targets for anti-aging treatments (Covarrubias et al., 2021). Research using genetically engineered animals affirms that loss of expression of the sirtuin genes Sirt1, Sirt3, Sirt6, or Sirt7 accelerates aging, while the overexpression of Sirt1, Sirt3, or Sirt6 potentially extends lifespans (Araki et al., 2015; Kanfi et al., 2012; Kawahara et al., 2009; Satoh et al., 2013; Vakhrusheva et al., 2008; Vazquez et al., 2016). SIRT7 is mainly localized in the nucleolus, and the levels gradually decrease during aging (Kiran et al., 2013). Accumulating evidence suggests SIRT7 as a stress sensor in physiological homeostasis (Sun et al., 2016; Yan et al., 2018; Yu et al., 2017). Exemplified by genotoxic stresses, SIRT7 is released from the nucleolus desuccinylates H3K122, deacetylates H4K18, and deglutarylates H4K91 to modulate chromatin confirmation, and facilitates the recruitment of 53BP1, enhancing the DNA damage response (Bao et al., 2019; Barber et al., 2012; Li et al., 2016; Vazquez et al., 2016). SIRT7 deacetylates and promotes the dephosphorylation of ATM after successful DNA repair (Tang et al., 2019), and its 5-fluorouracil-induced reduction promotes cancer radiosensitivity (Tang et al., 2017a). SIRT7 also cooperates with Myc activity to suppress ER stress (Shin et al., 2013), and the aging-related decline in SIRT7 compromises the regenerative capacity of hematopoietic stem cells by increasing mitochondrial protein-folding stress (Mohrin et al., 2015). Our data revealed SIRT7 as a novel epitranscriptional regulator of stress response.
NAT10 is mainly located in the nucleolus and is essential for nucleolus assembly during mitosis (Shen et al., 2009). NAT10 overexpression causes telomere shortening (Fu & Collins, 2007), and induces micronuclei formation in tumor cells, activating cGAS and leading to the production of the senescence-related secretory phenotype, a hallmark of cellular senescence (Cao et al., 2020). Recently, it has been reported that the increase in oxidative stress in elderly individuals increases cell ac4C levels and, thus, accelerates the degradation of tRNA, which leads to IL-1β production (Furman et al., 2017). In addition, pharmaceutical inhibition and genetic ablation of NAT10 can rescue progeroid features of cells and mouse models for Hutchinson-Gilford progeria syndrome, a premature aging disease (Balmus et al., 2018; Larrieu et al., 2014, 2018; Wilson, 2018). We found that NAT10 is unlikely to be a protein-deacetylating target of SIRT7, as comparable levels of pan-acetyl-lysine were observed in the anti-NAT10 immunoprecipitates in both the presence and absence of ectopic HA-SIRT7. Of note, another sirtuin member, SIRT1, deacetylates NAT10 to activate autophagy and enhance cell survival (Cai et al., 2017; Liu et al., 2018). It is, therefore, hypothesized that the ac4C modification plays a role in aging.
Our study provides the first evidence that SIRT7 is an eraser of RNA ac4C, and couples longevity-promoting sirtuins to epitranscriptomic regulation. Evidence points towards ac4C modification being involved in the regulation of aging, and this hypothesis merits further investigation.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Araki, S., Izumiya, Y., Rokutanda, T., Ianni, A., Hanatani, S., Kimura, Y., Onoue, Y., Senokuchi, T., Yoshizawa, T., Yasuda, O., Koitabashi, N., Kurabayashi, M., Braun, T., Bober, E., Yamagata, K., & Ogawa, H. (2015). Sirt7 contributes to myocardial tissue repair by maintaining transforming growth factor-beta signaling pathway. Circulation, 132, 1081–1093.
Arango, D., Sturgill, D., Alhusaini, N., Dillman, A. A., Sweet, T. J., Hanson, G., Hosogane, M., Sinclair, W. R., Nanan, K. K., Mandler, M. D., Fox, S. D., Zengeya, T. T., Andresson, T., Meier, J. L., Coller, J., & Oberdoerffer, S. (2018). Acetylation of cytidine in mRNA promotes translation efficiency. Cell, 175, 1872–18C6 e1824.
Balmus, G., Larrieu, D., Barros, A. C., Collins, C., Abrudan, M., Demir, M., Geisler, N. J., Lelliott, C. J., White, J. K., Karp, N. A., Atkinson, J., Kirton, A., Jacobsen, M., Clift, D., Rodriguez, R., Adams, D. J., & Jackson, S. P. (2018). Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nature Communications, 9, 1700.
Bao, X., Liu, Z., Zhang, W., Gladysz, K., Fung, Y. M. E., Tian, G., Xiong, Y., Wong, J. W. H., Yuen, K. W. Y., & Li, X. D. (2019). Glutarylation of histone H4 Lysine 91 regulates chromatin dynamics. Molecular Cell, 76, 660-675e669.
Barber, M. F., Michishita-Kioi, E., Xi, Y., Tasselli, L., Kioi, M., Moqtaderi, Z., Tennen, R. I., Paredes, S., Young, N. L., Chen, K., Struhl, K., Garcia, B. A., Gozani, O., Li, W., & Chua, K. F. (2012). SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature, 487, 114–118.
Cai, S., Liu, X., Zhang, C., Xing, B., & Du, X. (2017). Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochemical and Biophysical Research Communications, 483, 624–629.
Cao, Y., Yao, M., Wu, Y., Ma, N., Liu, H., & Zhang, B. (2020). N-acetyltransferase 10 promotes micronuclei formation to activate the senescence-associated secretory phenotype machinery in colorectal cancer cells. Translation Oncology. https://doi.org/10.1016/j.tranon.2020.100783
Chen, S., Seiler, J., Santiago-Reichelt, M., Felbel, K., Grummt, I., & Voit, R. (2013). Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Molecular Cell, 52, 303–313.
Chen, S., Blank, M. F., Iyer, A., Huang, B., Wang, L., Grummt, I., & Voit, R. (2016). SIRT7-dependent deacetylation of the U3–55k protein controls pre-rRNA processing. Nature Communications, 7, 10734.
Covarrubias, A. J., Perrone, R., Grozio, A., & Verdin, E. (2021). NAD(+) metabolism and its roles in cellular processes during ageing. Nature Reviews Molecular Cell Biology, 22, 119–141.
Dong, L., Yu, L., Li, H., Shi, L., Luo, Z., Zhao, H., Liu, Z., Yin, G., Yan, X., & Lin, Z. (2020). An NAD(+)-dependent deacetylase SIRT7 promotes HCC development through deacetylation of USP39. Iscience. https://doi.org/10.1016/j.isci.2020.101351
Ford, E., Voit, R., Liszt, G., Magin, C., Grummt, I., & Guarente, L. (2006). Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes & Development, 20, 1075–1080.
Fu, D., & Collins, K. (2007). Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Molecular Cell, 28, 773–785.
Furman, D., Chang, J., Lartigue, L., Bolen, C. R., Haddad, F., Gaudilliere, B., Ganio, E. A., Fragiadakis, G. K., Spitzer, M. H., Douchet, I., Daburon, S., Moreau, J.-F., Nolan, G. P., Blanco, P., Déchanet-Merville, J., Dekker, C. L., Jojic, V., Kuo, C. J., Davis, M. M., & Faustin, B. (2017). Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nature Medicine, 23, 174–184.
Ianni, A., Kumari, P., Tarighi, S., Simonet, N. G., Popescu, D., Guenther, S., Holper, S., Schmidt, A., Smolka, C., Yue, S., Krüger, M., Fiorillo, C., Vaquero, A., Bober, E., Krüger, T. B., Fiorillo, C., Vaquero, A., Bober, E., & Braun, T. (2021). SIRT7-dependent deacetylation of NPM promotes p53 stabilization following UV-induced genotoxic stress. Proceedings of the National Academy of Sciences. https://doi.org/10.1073/pnas.2015339118
Ito, S., Akamatsu, Y., Noma, A., Kimura, S., Miyauchi, K., Ikeuchi, Y., Suzuki, T., & Suzuki, T. (2014a). A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. Journal of Biological Chemistry, 289, 26201–26212.
Ito, S., Horikawa, S., Suzuki, T., Kawauchi, H., Tanaka, Y., Suzuki, T., & Suzuki, T. (2014b). Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). Journal of Biological Chemistry, 289, 35724–35730.
Jin, G., Xu, M., Zou, M., & Duan, S. (2020). The processing, gene regulation, biological functions, and clinical relevance of N4-acetylcytidine on RNA: A systematic review. Molecular Therapy Nucleic Acids, 20, 13–24.
Kanfi, Y., Naiman, S., Amir, G., Peshti, V., Zinman, G., Nahum, L., Bar-Joseph, Z., & Cohen, H. Y. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature, 483, 218–221.
Kawahara, T. L., Michishita, E., Adler, A. S., Damian, M., Berber, E., Lin, M., McCord, R. A., Ongaigui, K. C., Boxer, L. D., Chang, H. Y., & Chua, K. F. (2009). SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell, 136, 62–74.
Kiran, S., Chatterjee, N., Singh, S., Kaul, S. C., Wadhwa, R., & Ramakrishna, G. (2013). Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS Journal, 280, 3451–3466.
Kowalski, S., Yamane, T., & Fresco, J. R. (1971). Nucleotide sequence of the “denaturable” leucine transfer RNA from yeast. Science, 172, 385–387.
Larrieu, D., Britton, S., Demir, M., Rodriguez, R., & Jackson, S. P. (2014). Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science, 344, 527–532.
Larrieu, D., Vire, E., Robson, S., Breusegem, S. Y., Kouzarides, T., & Jackson, S. P. (2018). Inhibition of the acetyltransferase NAT10 normalizes progeric and aging cells by rebalancing the transportin-1 nuclear import pathway. Science Signaling. https://doi.org/10.1126/scisignal.aar5401
Lee, N., Kim, D. K., Kim, E. S., Park, S. J., Kwon, J. H., Shin, J., Park, S. M., Moon, Y. H., Wang, H. J., Gho, Y. S., & Choi, K. Y. (2014). Comparative interactomes of SIRT6 and SIRT7: Implication of functional links to aging. Proteomics, 14, 1610–1622.
Li, L., Shi, L., Yang, S., Yan, R., Zhang, D., Yang, J., He, L., Li, W., Yi, X., Sun, L., Liang, J., Cheng, Z., Shi, L., Shang, Y., & Yu, W. (2016). SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nature Communications, 7, 12235.
Li, G., Tang, X., Zhang, S., Jin, M., Wang, M., Deng, Z., Liu, Z., Qian, M., Shi, W., Wang, Z., Xie, H., Li, J., & Liu, B. (2020). SIRT7 activates quiescent hair follicle stem cells to ensure hair growth in mice. The EMBO Journal. https://doi.org/10.15252/embj.2019104365
Liu, X., Cai, S., Zhang, C., Liu, Z., Luo, J., Xing, B., & Du, X. (2018). Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Research, 46, 9601–9616.
Liu, Z., Qian, M., Tang, X., Hu, W., Sun, S., Li, G., Zhang, S., Meng, F., Cao, X., Sun, J., Xu, C., Tan, B., Pang, Q., Zhao, B., Wang, Z., Guan, Y., Ruan, X., & Liu, B. (2019). SIRT7 couples light-driven body temperature cues to hepatic circadian phase coherence and gluconeogenesis. Nature Metabolism, 1, 1141–1156.
Lu, Y. F., Xu, X. P., Lu, X. P., Zhu, Q., Liu, G., Bao, Y. T., Wen, H., Li, Y. L., Gu, W., & Zhu, W. G. (2020). SIRT7 activates p53 by enhancing PCAF-mediated MDM2 degradation to arrest the cell cycle. Oncogene, 39, 4650–4665.
Meng, F., Qian, M., Peng, B., Peng, L., Wang, X., Zheng, K., Liu, Z., Tang, X., Zhang, S., Sun, S., Cao, X., Pang, Q., Zhao, B., Ma, W., Songyang, Z., Xu, B., Zhu, W.-G., Xu, X., & Liu, B. (2020). Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. eLife. https://doi.org/10.7554/eLife.55828
Mitra, N., & Dey, S. (2020). Biochemical characterization of mono ADP ribosyl transferase activity of human sirtuin SIRT7 and its regulation. Archives of Biochemistry and Biophysics. https://doi.org/10.1016/j.abb.2019.108226
Mohrin, M., Shin, J., Liu, Y., Brown, K., Luo, H., Xi, Y., Haynes, C. M., & Chen, D. (2015). Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science, 347, 1374–1377.
O’Callaghan, C., & Vassilopoulos, A. (2017). Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell, 16, 1208–1218.
Roundtree, I. A., Evans, M. E., Pan, T., & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169, 1187–1200.
Ryu, D., Jo, Y. S., Lo Sasso, G., Stein, S., Zhang, H., Perino, A., Lee, J. U., Zeviani, M., Romand, R., Hottiger, M. O., Schoonjans, K., & Auwerx, J. (2014). A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metabolism, 20, 856–869.
Sas-Chen, A., Thomas, J. M., Matzov, D., Taoka, M., Nance, K. D., Nir, R., Bryson, K. M., Shachar, R., Liman, G. L. S., Burkhart, B. W., Gamage, S. T., Nobe, Y., Briney, C. A., Levy, M. J., Fuchs, R. T., Robb, G. B., Hartmann, J., Sharma, S., Lin, Q., … Schwartz, S. (2020). Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature, 583, 638–643.
Satoh, A., Brace, C. S., Rensing, N., Cliften, P., Wozniak, D. F., Herzog, E. D., Yamada, K. A., & Imai, S. (2013). Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metabolism, 18, 416–430.
Sharma, S., Langhendries, J. L., Watzinger, P., Kotter, P., Entian, K. D., & Lafontaine, D. L. (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Research, 43, 2242–2258.
Shen, Q., Zheng, X., McNutt, M. A., Guang, L., Sun, Y., Wang, J., Gong, Y., Hou, L., & Zhang, B. (2009). NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Experimental Cell Research, 315, 1653–1667.
Shin, J., He, M., Liu, Y., Paredes, S., Villanova, L., Brown, K., Qiu, X., Nabavi, N., Mohrin, M., Wojnoonski, K., Li, P., Cheng, H.-L., Murphy, A. J., Valenzuela, D. M., Luo, H., Kapahi, P., Krauss, R., Mostoslavsky, R., Yancopoulos, G. D., … Chen, D. (2013). SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Reports, 5, 654–665.
Sinclair, W. R., Arango, D., Shrimp, J. H., Zengeya, T. T., Thomas, J. M., Montgomery, D. C., Fox, S. D., Andresson, T., Oberdoerffer, S., & Meier, J. L. (2017). Profiling cytidine acetylation with specific affinity and reactivity. ACS Chemical Biology, 12, 2922–2926.
Sun, L., Fan, G., Shan, P., Qiu, X., Dong, S., Liao, L., Yu, C., Wang, T., Gu, X., Li, Q., Song, X., Cao, L., Li, X., Cui, Y., Zhang, S., & Wang, C. (2016). Regulation of energy homeostasis by the ubiquitin-independent REGgamma proteasome. Nature Communications, 7, 12497.
Tang, B. L. (2015). SIRT7 and hepatic lipid metabolism. Frontiers in Cell and Developmental Biology, 3, 1.
Tang, M., Lu, X., Zhang, C., Du, C., Cao, L., Hou, T., Li, Z., Tu, B., Cao, Z., Li, Y., Chen, Y., Jiang, L., Wang, H., Wang, L., Liu, B., Xu, X., Luo, J., Wang, J., Gu, J., … Zhu, W.-G. (2017a). Downregulation of SIRT7 by 5-fluorouracil induces radiosensitivity in human colorectal cancer. Theranostics, 7, 1346–1359.
Tang, X., Shi, L., Xie, N., Liu, Z., Qian, M., Meng, F., Xu, Q., Zhou, M., Cao, X., Zhu, W. G., & Liu, B. (2017b). SIRT7 antagonizes TGF-beta signaling and inhibits breast cancer metastasis. Nature Communications, 8, 318.
Tang, M., Li, Z., Zhang, C., Lu, X., Tu, B., Cao, Z., Li, Y., Chen, Y., Jiang, L., Wang, H., Wang, L., Wang, J., Liu, B., Xu, X., Wang, H., & Zhu, W.-G. (2019). SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Science Advances. https://doi.org/10.1126/sciadv.aav1118
Thalalla Gamage, S., Sas-Chen, A., Schwartz, S., & Meier, J. L. (2021). Quantitative nucleotide resolution profiling of RNA cytidine acetylation by ac4C-seq. Nature Protocols, 16, 2286–2307.
Tong, Z., Wang, M., Wang, Y., Kim, D. D., Grenier, J. K., Cao, J., Sadhukhan, S., Hao, Q., & Lin, H. (2017). SIRT7 is an RNA-activated protein lysine deacylase. ACS Chemical Biology, 12, 300–310.
Tsai, Y. C., Greco, T. M., & Cristea, I. M. (2014). Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Molecular and Cellular Proteomics, 13, 73–83.
Vakhrusheva, O., Smolka, C., Gajawada, P., Kostin, S., Boettger, T., Kubin, T., Braun, T., & Bober, E. (2008). Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circulation Research, 102, 703–710.
Vazquez, B. N., Thackray, J. K., Simonet, N. G., Kane-Goldsmith, N., Martinez-Redondo, P., Nguyen, T., Bunting, S., Vaquero, A., Tischfield, J. A., & Serrano, L. (2016). SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO Journal, 35, 1488–1503.
Wang, W. W., Angulo-Ibanez, M., Lyu, J., Kurra, Y., Tong, Z., Wu, B., Zhang, L., Sharma, V., Zhou, J., Lin, H., Gao, Y. Q., Li, W., Chua, K. F., & Liu, W. R. (2019). A click chemistry approach reveals the chromatin-dependent histone H3K36 deacylase nature of SIRT7. Journal of the American Chemical Society, 141, 2462–2473.
Wiener, D., & Schwartz, S. (2021). The epitranscriptome beyond m(6)A. Nature Reviews Genetics, 22, 119–131.
Wilson, K. L. (2018). Nuclear import pathway key to rescuing dominant progerin phenotypes. Science Signaling. https://doi.org/10.1126/scisignal.aat9448
Yan, W. W., Liang, Y. L., Zhang, Q. X., Wang, D., Lei, M. Z., Qu, J., He, X. H., Lei, Q. Y., & Wang, Y. P. (2018). Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Reports. https://doi.org/10.15252/embr.201846377
Yoshizawa, T., Karim, M. F., Sato, Y., Senokuchi, T., Miyata, K., Fukuda, T., Go, C., Tasaki, M., Uchimura, K., Kadomatsu, T., Tian, Z., Smolka, C., Sawa, T., Takeya, M., Tomizawa, K., Ando, Y., Araki, E., Akaike, T., Braun, T., … Yamagata, K. (2014). SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metabolism, 19, 712–721.
Yu, J., Qin, B., Wu, F., Qin, S., Nowsheen, S., Shan, S., Zayas, J., Pei, H., Lou, Z., & Wang, L. (2017). Regulation of serine-threonine kinase Akt activation by NAD(+)-dependent deacetylase SIRT7. Cell Reports, 18, 1229–1240.
Zachau, H. G., Dütting, B., & Feldmann, H. (1966). The structures of two serine transfer ribonucleic acids. Hoppe-Seyler’s Zeitschrift Fur Physiologische Chemie, 347, 212–235.
Acknowledgements
The authors would like to thank Dr. Jessica Tamanini (Shenzhen University and ETediting) for editing the manuscript prior to submission.
Funding
This study was supported by grants from the National Natural Science Foundation of China (32090030, 91849208 and 82061160495), the National Key R&D Program of China (2017YFA0503900), and the Shenzhen Municipal Commission of Science and Technology Innovation (ZDSYS20190902093401689, KQJSCX20180328093403969 and JCYJ20180507182044945).
Author information
Chenzhong Xu, Jin Zhang and Jie Zhang have contributed equally.
Affiliations
Shenzhen Key Laboratory for Systemic Aging and Intervention (SAI), Guangdong Key Laboratory of Genome Stability and Human Disease Prevention, National Engineering Research Center for Biotechnology (Shenzhen), International Cancer Center, School of Basic Medical Sciences , Shenzhen University, Shenzhen, 518055, China
Chenzhong Xu, Jin Zhang & Baohua Liu
Shenzhen University (SZU)-Friedrich Schiller University (FSU) Jena Joint PhD Program, Shenzhen University, Shenzhen, 518055, China
Jie Zhang
Shenzhen Bay Laboratory, Shenzhen, 518055, China
Baohua Liu
Corresponding author
Correspondence to Baohua Liu.
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, C., Zhang, J., Zhang, J. et al. SIRT7 is a deacetylase of N4-acetylcytidine on ribosomal RNA. GENOME INSTAB. DIS. 2, 253–260 (2021). https://doi.org/10.1007/s42764-021-00046-x
Received20 May 2021
Revised05 July 2021
Accepted06 July 2021
Published14 July 2021
Issue DateAugust 2021
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable linkKeywords
RNA N4-acetylcytidine (ac4C)
SIRT7
NAT10








用户登录
还没有账号?
立即注册