Biological function and regulation of histone 4 lysine 20 methylation in DNA damage response
Review Article
Open Access
Genome Instability & Disease 3, 33–46 (2022)
Abstract
Cells are often under attack from various DNA-damaging agents. Accurate repair is required to protect cells from the genome instability induced by DNA lesions. DNA damage response (DDR) signaling involves sensitizing, transmitting, and repairing different types of damage within chromatin complexes. Chromatin is a highly ordered complex packed with repeating units of nucleosomes and linker DNA sequences. Chromatin structure, gene transcription, and various biological processes are regulated by histone post-translational modifications (PTMs), including acetylation, methylation, phosphorylation, and ubiquitylation. Of these, the involvement of lysine methylation, regulated by numerous lysine methyltransferases and demethylases, in the DDR has been extensively explored. In particular, histone 4 lysine 20 methylation is one of the most essential histone PTMs for biological processes and ensures genome integrity. In this review, we summarize the dynamics and modulations of histone lysine methylation during the DDR. We also comprehensively describe the functions, mechanisms, and regulation of H4K20 methylation and its modifying enzymes in response to DNA damage.
Introduction
DNA in eukaryotic cells is continually being damaged in an abundance of ways. This damage usually includes DNA single-strand breaks (SSBs), DNA double-strand breaks (DSBs), adducts, intrastrand and interstrand cross-links, and insertion/deletion mismatches. Among them, DSBs are considered the most cytotoxic type of damage because they induce genomic instability. Ionizing radiation (IR), ultraviolet light, and various chemical agents represent commonly exogenous sources of DNA damage. Deregulated DNA replication can also lead to DNA damage, and the disruption of replication forks has been shown to cause severe lesions, including DSBs (Jackson & Bartek, 2009; Lord & Ashworth, 2012). To preserve genomic integrity, mammalian cells repair damaged DNA via at least nine distinct pathways (Ceccaldi et al., 2016; Clauson et al., 2013; Kennedy & D'Andrea, 2006; Kramara et al., 2018). For example, IR-induced DSBs are mainly repaired by non-homologous end-joining (NHEJ) and homologous recombination (HR). Each repair pathway is detected and activated by different repair factors to form a precise network that ensures genome stability.
The nucleosome is the basic structural unit consisting of two copies of the histone proteins, H4, H3, H2A, and H2B, and is wrapped in 147 bp of DNA nucleotides. They are further packaged into a 30-nm fiber to form the chromatosome core particle consisting of a linker histone (H1) bound to the nucleosome with 10 bp of DNA. Histone tails are susceptible to a variety of covalent post-translational modifications (PTMs), including methylation, acetylation, ubiquitination, and phosphorylation. The PTMs of the histone tails tightly control many biological processes by regulating chromatin structure and function (Van & Santos, 2018). The second most common histone modification linked to the DNA damage response (DDR) is histone methylation, which is under the control of histone methyltransferase (HMT) and the histone demethylases (HDMs) (Chen & Zhu, 2016). Both the lysine and arginine residues of histones can be methylated, and histone methylation presents as three forms: mono-(me1), di-(me2), and rei-(me3). It has been demonstrated that histone methylation, particularly lysine methylation, plays a crucial role in the response to DNA damage (Uckelmann & Sixma, 2017; J. Zhang et al., 2021). Methylations of lysine are modulated by lysine methyltransferases (KMTs) and lysine demethylases (KDMs). While the role of histone acetylation in the DDR has been extensively studied (Li & Zhu, 2014; Li et al., 2020), the investigation of histone methylation is still at a relatively early stage. Therefore, elucidating the mechanisms and functions of histone lysine methylation will be helpful for developing novel therapeutic strategies for DNA-damage-related diseases, including aging and cancer.
The common lysine methylation sites are residues 4, 9, 27, 36, and 79 of histone H3 and lysine 16 and 20 of histone H4. Genome-wide localization analyses indicate that each residue and degree-specific methylation has a distinct distribution pattern, indicating their unique functions during diverse cellular processes. Lysine 20 is a major methylated lysine residue on the histone H4 tail. Each degree of methylation at this residue has distinct regulation properties and shares similar functions during the DDR. For example, H4K20me1 regulates the cell cycle through condensin II, H4K20me2 negatively correlates with H4K16ac, and H4K20me3 is highly enriched in heterochromatin. Moreover, all three forms of H4K20 methylation are reported to contribute to p53-binding protein 1 (53BP1) recruitment (Tan et al., 2011). A better understanding of the mechanisms that regulate H4K20 methylation will enable us to gain deeper insights into how different degrees of methylation are controlled and what cellular machinery requires this modification. In this review, we summarize the critical roles of histone lysine methylation, especially H4K20 methylation, and the relevant modulating enzymes in the DDR.
Histone lysine methylation
Histone methylation has been recognized as being a PTM since 1964 (Murray, 1964). Since this time, methylation has been identified on various lysine residues on histone H3 and H4 tails, including H3K4me1/2, H3K9me2/3, H3K27me2/3, H3K36me1/2/3, H3K79me1/2/3, H4K16me1, and H4K20me1/2/3 (Husmann & Gozani, 2019). Other basic residues, e.g., H1.4 and H2AX, have been shown to be methylated in the nucleosome (Black et al., 2012) (Fig. 1). Histone methylation plays an essential role in the DDR, chromatin structure, gene transcription, and cell cycle regulation. To date, many histone methylation sites have been shown to be related to the DDR in different ways. Histone methylation can provide high-affinity binding sites or platforms for repair factor recruitment (Chen & Zhu, 2016). In addition, DNA-histone interactions in the chromatin structure are mediated by the attraction between the negatively charged DNA backbone and positively charged lysine and arginine residues of the histones. Although histone methylation cannot directly change the charge of histones, it is involved in crosstalk with other histone modifications, such as acetylation, to regulate the compaction of chromatin (Cao et al., Cao, Shen, et al., 2016, Cao, Wei, et al., 2016). To date, although almost all identified histone methylation sites reportedly participate in the DDR, deeper investigation is required into whether and how crosstalk among different sites affects DNA repair.
Fig. 1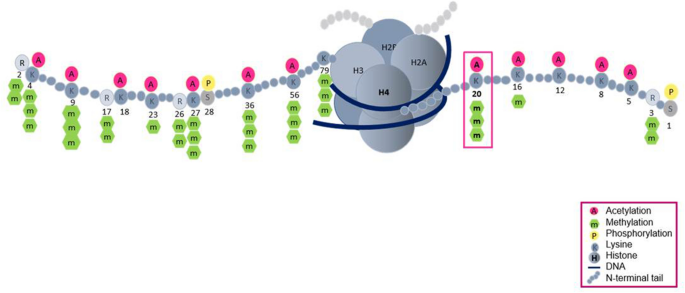
Overview of histone modifications. Representative modification sites are shown. Numbers indicate lysine or arginine residues on histones H3 and H4
Full size imageH3K4 is a major methylation site of histones and is often associated with transcriptionally active genes (Santos-Rosa et al., 2002). At sites of DNA damage, H3K4me3 regulates NHEJ efficiency and S-phase transition in yeast cells (Faucher & Wellinger, 2010). The methylation of H3K9 is tightly linked to stable inheritance of the heterochromatic state (Bannister et al., 2001; Rea et al., 2000). H3K9me3 interacts with the chromo domain of Drosophila heterochromatin protein 1 (HP1) to regulate chromatin compaction during the DDR (Jacobs & Khorasanizadeh, 2002). Loss of H3K9me3 at DSBs leads to defective repair and increased radiosensitivity (Ayrapetov et al., 2014). H3K14 methylation, the latest methylation site discovered on H3, negatively correlates with H3K14 acetylation to repress host gene expression (Rolando et al., 2013). Recently, H3K14me3 is reported to respond to replication stress by enhancing ataxia-telangiectasia-mutated-and-Rad3-related kinase (ATR) activation (Zhu et al., 2021). H3K27 methylation is an established gene-silencing marker (Cao et al., 2002; Margueron & Reinberg, 2011). H3K27me2/3 facilitates NHEJ efficiency by triggering the chromatin accumulation of Fanconi anemia complementation group D2, a central player in the cell’s choice of DNA repair pathway (Zhang et al., 2018). H3K36 methylation is associated with the elongating form of Pol II (Wagner & Carpenter, 2012). H3K36me2 recruits and stabilizes DNA repair components, including Nijmegen breakage syndrome protein 1 and Ku70, at DSBs (Fnu et al., 2011). H3K36me3 promotes HR repair by facilitating DNA end resection (Pfister et al., 2014). H3K79 methylation participates in active transcription and the DDR (Huyen et al., 2004; Mehta et al., 2016). H3K79me3 binds to 53BP1 to regulate the higher-order chromatin structure during the DDR (Huyen et al., 2004). H4K16me1 was originally suggested following a mass spectrometry study a decade ago, its biological function in the DDR has only recently been clarified (Lu et al., 2019; Tan et al., 2011). H4K16me1 can cooperate with H4K20me2 to facilitate 53BP1 recruitment and NHEJ mediated DNA repair (Lu et al., 2019). H4K20 methylation is one of the most thoroughly studied histone modifications: it has diverse functions and is regulated by various histone modifiers (Jorgensen et al., 2013). The role H4K20 methylation in DDR will be discussed in detail in this review. The first mammalian H1 methylation site identified was H1.4K26. The methylation of H1.4K26 provides a recognition surface for the chromatin-binding of HP1 and lethal 3 malignant brain tumor 1 (L3MBTL1) (Trojer et al., 2009; Walport et al., 2018). Although most common methylation sites are reported to be involved in DNA repair efficiency, the detailed mechanisms underlying how specific site contributes to DDR require further investigations. For example, although H3K4me3 are shown to be important for responses to DNA damaging agents, understanding the contribution of H3K4 methylation and its methyltransferases in repair factors recruitment needs further studies. In addition, several other identified methylation sites exist, but their biological modulation, especially about DDR, remains unclear. For example, little is known about the methylation of H4 K5, K8, or K12 catalyzed by yeast Set5, except for its role in cell growth and stress responses (Green et al., 2012).
Histone lysine methyltransferases and the DNA damage response
HMTs are enzymes that transfer the methyl groups from S-adenoidal-L-methionine (SAM) to histone proteins (Fig. 2). HMTs are divided into three groups, SET-domain-containing proteins, DOT1-like proteins, and protein arginine N-methyltransferases (PRMTs). The first two families are lysine KMTs, whereas the PRMT family has been shown to methylate arginine. In mammalian cells, PRMTs consist of PRMT1 to PRMT9, whereas KMTs encompass KMT1 to KMT9 (Greer & Shi, 2012). The roles of KMTs vary with their histone substrates or interacting partners, and many KMTs are thought to participate in the DDR (Biggar & Li, 2015) (Table 1).
Fig. 2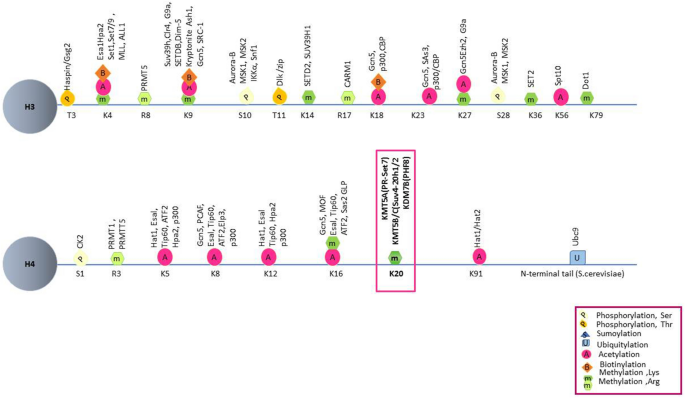
Histone modifications and modifiers. Each residue with known modifiers on histones H3 and H4 are shown for each specific site
Full size imageTable 1 Histone lysine-specific methyltransferases and DNA damage repairFull size tableKMT1A (SUV39H1) is a methyltransferase of H3K9 methylation and is linked to the organization of higher-order chromatin and the recruitment of HP1 to DSBs (Ayrapetov et al., 2014; Rea et al., 2000; Tu et al., 2020). KMT1B (SUV39H2) is a methyltransferase of H2AXK134me2 (Sone et al., 2014), which regulates γ-H2AX levels during the DDR (Sone et al., 2014). KMT1C (G9a, EHMT2) methylates H3K9me1/2 and H1.4K26me1/2 to provide a recognition surface for the chromatin-binding of replication protein A (RPA), HP1, and L3MBTL1 (Trojer et al., 2009; Yang et al., 2017). KMT1D (GLP, EHMT1), the methyltransferase of H3K9me1/2 and H4K16me1, facilitates 53BP1 recruitment in the NHEJ pathway (Lu et al., 2019; Takahashi et al., 2012). KMT1E (SETDB1) methylates H3K9me3, which promotes 53BP1 reposition at damaged sites (Alagoz et al., 2015). The methyltransferase KMT3A (SETD2) catalyzes H3K14me3 and H3K36me3 to facilitate RPA complex loading onto chromatin and HR repair, respectively (Pfister et al., 2014; Zhu et al., 2021). KMT4 (DOT1L) methylates H4K79me1/2/3 to recruit 53BP1 to DSBs (Huyen et al., 2004). KMT5A (PR-Set7, SET8, SETD8) is responsible for H4K20me1, which is involved in DNA damage accumulation, 53BP1 recruitment, S-phase checkpoint, and ATR dependent cell cycle arrest (Beck et al., 2012; Li et al., 2016; Tuzon et al., 2014). KMT5B (SUV4-20H1) catalyzes H4K20me2 to regulate the licensing and activation of early replication origins and maintains replication timing (Long et al., 2020). KMT5C (SUV4-20H2) catalyzes H4K20me3, allowing it to function in repetitive element silencing (Fodor et al., 2010), and the elimination of KMT5B/C proteins causes defected S-phase entry, whereas loss of KMT5A leads to fatal consequences (Schotta et al., 2004). KMT6A (EZH2) is responsible for H3K27me3 and accumulates at actively transcribes gene promoters during DDR (Karakashev et al., 2020; Kuser-Abali et al., 2018). KMT7 (SET7/9) demethylates P53 and SUV39H1 to facilitate an efficient DDR (Liu et al., 2011; Wang et al., 2013), KMT8A (PRDM2) promotes recruitment of BReast-CAncer susceptibility gene 1 (BRCA1), but not 53BP1, to contribute to chromatin condensation (Khurana et al., 2014). SETMA (Metnase) methylates H3K4 and H3K36, which are associated with chromatin opening (Lee et al., 2005). Although many studies confirmed the recruitment of different KTMs, their specific methylation site are not increased. For example, KMT6 accumulates at DSB site, while the increase of H3K27me3 is not detected during DNA damage repair (Zhang et al., 2021). Other substrates, including histones and non-histones, might be involved in this progress.
Histone demethylases and the DNA damage response
HDMs, which are responsible for methyl group removal, are divided into histone arginine demethylases and KDMs (Chang et al., 2019; Dimitrova et al. 2015; Shi et al., 2003, 2004). KDMs are further divided into two groups: the first is the mono-amine oxidases, which use flavin adenine dinucleotide (FAD) as a cofactor to oxidize methyl groups and hydrolyze them into formaldehyde, and the second is the Jumonji-C (JmjC) class, which uses Fe(II) and 2-oxoglutarate (2-OG or α-ketoglutarate) as cofactors to hydroxylate the methyl groups via a free-radical mechanism (Jambhekar et al. 2017). Many KDMs are reported to participate in the DDR (Table 2).
Table 2 Histone lysine-specific demethylases and DNA damage repairFull size tableKDM1A (LSD1) specifically demethylates H3K4 methylation, which is linked to active transcription (Shi et al., 2004); KDM1A lies downstream of RING finger protein 168 (RNF168) but upstream of 53BP1 and BRCA1 in the DDR (Mosammaparast et al., 2013). KDM2A (JHDM1A, FBXL11), the demethylase for H3K36me3, acts as a substrate of Ataxia telangiectasia mutated kinase (ATM) to modulate the recruitment of the meiotic recombination 11 ( MRE11) complex to DNA-damage sites (Cao et al., Cao, Shen, et al., 2016, Cao, Wei, et al., 2016). KDM2B (JHDM1B, FBXL10) is responsible for the timely dissociation of proliferating cell nuclear antigen (PCNA) from chromatin, allowing for efficient DNA replication (Kang et al., 2020). Four proteins belong to the KDM4, also known as JMJD2, encompasses four subfamilies, including KDM4A (JMJD2A), KDM4B (JMJD2B), KDM4C (JMJD2C), and KDM4D (JMJD2D), all of which have a JmjC domain. KDM4 demethylates H3K9me2/3, H3K36me2/3, and H1.4K26 to regulate gene expression and chromatin structure (Lee et al., 2020). The degradation of KDM4A and KDM4B restores the formation of 53BP1 foci in RING finger protein 8 (RNF8)- and RNF168-deficient cells (Mallette et al., 2012), and KDM4D knockdown disrupts the DNA-damage-induced association between ATM, RAD51, 53BP1, and chromatin (Khoury-Haddad et al., 2014). KDM5A-dependent H3K4me3 demethylation is required for Zinc finger and MYND (Myeloid, Nervy, and DEAF-1) domain containing 8 (ZMYND8)–Nucleosome remodeling and histone deacetylation (NuRD) recruitment to DNA damage sites (Gong et al., 2017). KDM5B contributes to the recruitment of Ku70 and BRCA1 to facilitate efficient DSB repair (Li et al., 2014). The KDM7 subfamily consists of KDM7A (KIAA1718), KDM7B (plant homeodomain finger-containing protein 8 [PHF8]), and KDM7C (PHF2). KDM7C has recently been found to control the formation of 53BP1 and BRCA1 (Alonso-de Vega et al., 2020). Although many KDMs are reported to accumulate at DSB sites, our knowledge of most KDMs is largely lacking. The functions of two demethylases those are responsible for different forms of the same methylation site could be opposite. For example, deficiency of KDM1A, the demethylase of H3K4me2, increased the HR repair efficiency, whereas loss of KDM5A/B, the demethylases of H3K4me2/3, impaired HR (Li et al., 2014; Mosammaparast et al., 2013). This contradiction might indicate that additional non-histone sustrates are involved.
Histone H4 lysine 20 methylation
H4K20 was one of the earliest described histone modifications was discovered in 1969 and forms the majority of lysine residue methylation on the H4 N-terminal tail (Fang et al., 2002; Schotta et al., 2004). Studies have shown that H4K20 must first be mono-methylated then di- and tri-methylated (Weirich et al., 2016; Wu et al., 2013). Several different methyltransferases and demethylases have been identified that regulate H4K20 methylation in mammals (Beck et al., 2012; Oda et al., 2010) (Fig. 3). KMT5A was previously recognized as the only mono-methyltransferase, whereas H4K20me2 and H4K20me3 are catalyzed by KMT5B/KMT5C (Nishioka et al., 2002; Schotta et al., 2008). KDM7B demethylates H4K20me1, while the demethylase for H4K2/3 is still largely unclear (Qi et al., 2010). Of note, KDM7B may not be the sole histone demethylase for H4K20me1, as KDM7B is not present in yeast (Klose et al., 2006), while H4K20me1 is (Edwards et al., 2011; Sanders et al., 2004). Consistent with this notion, using high-content cell-based screening of 2500 nuclear proteins, hHR23A was identified as a demethylase for H4K20me1/2/3 (X. Cao et al., 2020). In addition, DPY-21 was recently also found to be an H4K20me2 demethylase (Brejc et al., 2017). Since the expression and activity of KMT5A are regulated by the cell cycle, the methylation state of H4K20 exhibits dynamic change (Liu et al., 2010) (Fig. 4). H4K20me1 is in decline during the G1 phase, leading to the accumulation of the unmodified form of H4K20 at the beginning of the S phase. H4K20me1 accumulates at the G2/M phase and then is gradually converted to H4K20me2/3 (Oda et al., 2009; Pesavento et al., 2008). H4K20 methylation plays an essential role in chromatin compaction, genome integrity, DNA damage repair, and DNA replication (Jorgensen et al., 2013). H4K20me1 and H4K20me2 are often associated with DNA damage repair and DNA replication after exposure to DNA damaging agents, whereas H4K20me3 is associated with the silencing of the heterochromatic state (Jorgensen et al., 2013).
Fig. 3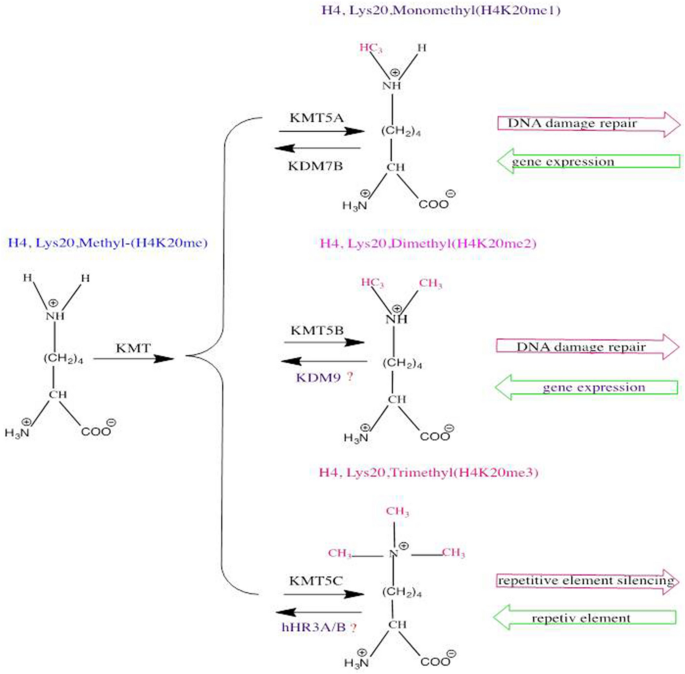
Schematic diagram of H4K20 methylation. The different methylation states of H4K20 with the various mammalian enzymes that perform these post-translational modifications are shown
Full size imageFig. 4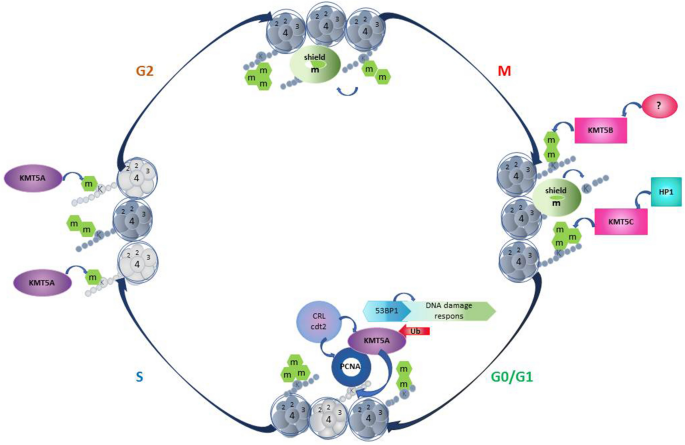
Diagram of H4K20 methylation during cell cycle regulation. The dynamics of H4K20me1/2/3 and established enzymes during cell cycle progression are depicted
Full size imageHistone H4 lysine 20 mono-methylation
The fundamental role of H4K20me1 in DNA repair comes from analyses of the first identified mono-methyltransferase, KTM5A (Lu et al., 2021). Deletion of KTM5A results in lethality in mice because of the huge numbers of DSBs (Oda et al., 2009). In mammalian cells, RNA interference of KTM5A expression leads to increased γH2AX foci, recruitment of DNA repair proteins, and activation of DNA damage checkpoint (Paulsen et al., 2009; Sakaguchi & Steward, 2007). The significant role of KTM5A in DNA damage may partially lie in its regulation of the cell cycle, as it was shown that defects in KTM5A and H4K20me1 demethylation result in a delay in G1-S transition (W. Liu et al., 2010). In addition, loss of KTM5A causes DNA damage, specifically during DNA replication, which is abrogated by the co-depletion of RAD51, a critical HR repair factor. H4K20me1 functions in DNA damage in direct and indirect ways. On one hand, KMT5A-mediated H4K20me1 directly regulates chromatin structure, without other cofactors, to contribute to DNA repair. On the other hand, H4K20me1 serves as a platform for cooperation with its binders, including 53BP1, L3MBTL1, condensin II, and PCNA.
BRCA1 and 53BP1 play essential roles in DNA repair with HR and NHEJ, respectively (Shibata, 2017). 53BP1 is the DNA damage checkpoint protein required for an efficient NHEJ pathway (FitzGerald et al., 2009), and in the absence of 53BP1, BRCA1 promotes repair through the HR pathway (Shibata, 2017). RNF8 and RNF168 cause the ubiquitination of H2AK15 via its ubiquitination dependent recruitment (UDR) motif; then H2AK15ub is recognized by 53BP1 and facilitates 53BP1 foci formation in the NHEJ pathway (Fradet-Turcotte et al., 2013) (Fig. 5).
Fig. 5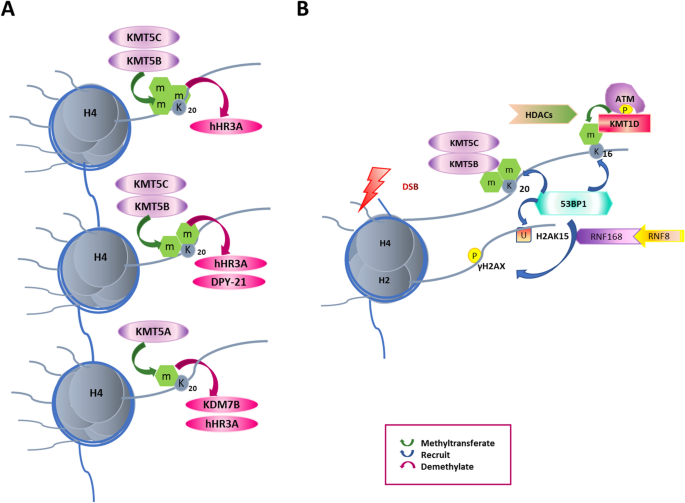
53BP1 is highly correlated with H4K20 methylation during DDR. A Essential methyltransferases of H4K20me/1/2/3 are known as KMT5A/B/C. KDM4A and KDM7B regulate H4K20 di/tri-demethylation. The crosstalk between H4K20 and H4K16 is also indicated. B 53BP1 binds to H4K20me/1/2/3 to facilitate DDR through the NHEJ pathway. After DSBs, ATM is auto-phosphorylated and further phosphorylates H2AX. RNF8/RNF168 ubiquitinates H2AK15 and disturbs the association of 53BP1 and H4K20 methylation
Full size imageH4K20me1 is linked to 53BP1 in the DDR, an association that was first identified in Schizosaccharomyces pombe. Crb9, the homolog of 53BP1, binds to Set9-catalyzed H4K20me1 for its recruitment to DNA damage sites (Sanders et al., 2004), an interaction that is also present in mammalian cells (Botuyan et al., 2006). A further study found that the tandem Tudor domain of 53BP1 is conserved and required for its recruitment. Knockdown of KMT5A inhibits 53BP1 recruitment to DNA damage sites and blocks checkpoint signaling, validating the idea that H4K20me1 contributes to 53BP1 recruitment (Oda et al., 2010). Interestingly, several studies demonstrated that Cullin ring-finger ubiquitin ligase 4 (CRL4, Cdt2) degrades chromatin-bound KMT5A via the PCNA-interacting motif (PIP-box). However, there is no increase in H4K20 methylation or recruitment of 53BP1 before KMT5A degradation (Abbas et al., 2010; Centore et al., 2010; Jorgensen et al., 2007). In addition, 53BP1 transfers to damaged sites even in the absence of KTM5A, indicating that there are other proteins and histone modifications involved in the 53BP1 recruitment platform and that KMT5A has other functions in addition to 53BP1 recruitment (Jorgensen et al., 2007; Tardat et al., 2007). In support of this notion, the ectopic expression of a non-degradable form of KMT5A led to chromatin condensation, activation of the DDR, and cell cycle progression failure (Beck et al., 2012).
Another factor that is recruited by H4K20me1 is L3MBTL1 (Trojer et al., 2007), which directly binds components of the replication machinery, potentially linking H4K20me1 with genome instability (Gurvich et al., 2010). Condensin II was also discovered to recognize H4K20me1 through its N-CAPD3 and N-CAPG2 subunits via two Huntingtin, elongation factor 3, protein phosphatase 2A, and lipid kinase TOR domains, and it is involved in mitotic progression (Liu et al., 2010).
Histone H4 lysine20 di/tri-methylation
H4K20me2 is an extremely abundant marker exhibited by more than 80% of nucleosomes, whereas H4K20me1 and H4K20me3 are present at comparably low levels. KMT5B and KMT5C are responsible for most of the di- and tri-methylation of H4K20, respectively (Schotta et al., 2004). KMT5B and KMT5C require SAM as a co-factor in the transfer of a methyl group to proteins, lipids, or nucleic acid substrates (Schotta et al., 2004; Southall et al., 2014; Wu et al., 2013). The KMT5B/C enzymes are characteristics of the DDR; they maintain telomere length and regulate heterochromatin compaction, and therefore are linked to genome instability (Schotta et al., 2008). In human cells, an inhibitor of KMT5B/C (A-196) has been shown to significantly impact genome integrity (Bromberg et al., 2017).
H4K20me2 has been observed to further accumulate at DNA-damage sites and to play a role in DNA-damage repair mainly by serving as a 53BP1-binding platform (Pei et al., 2011). In agreement with this, KMT5A was reportedly insufficient for 53BP1 recruitment. The de novo H4K20me1 facilitates KTM5B/C recruitment and catalyzes H4K20me2/3 to regulate 53BP1 binding (Tuzon et al., 2014). Actually, peptide-affinity studies suggested that 53BP1 has a higher affinity for H4K20me2 peptide than for H4K20me1 peptide (Schotta et al., 2004). KTM5B/C double-knockout cells showed significantly delayed 53BP1 foci formation (Schotta et al., 2008). In addition to KTM5B/C, multiple myeloma SET domain-containing protein (MMSET), the primary methyltransferase for H3K36me, has also been identified as a methyltransferase for H4K20me2/3, but not H4K20me1. The downregulation of MMSET significantly decreases the accumulation of 53BP1 (Pei et al., 2011). Besides methyltransferases, several other proteins containing a Tudor domain bind to H4K20me2 to regulate 53BP1 recruitment in response to DNA damage. KDM4A competes with 53BP1 for binding to H4K20me2 and facilitates 53BP1 movement to sites of damage; KDM4A is degraded in an RNF8- and RNF168-dependent manner after DNA damage (Mallette et al., 2012). In contrast to human L3MBTL1, which mainly binds to H4K20me1, dl(3)MBT binds to all three methylated H4K20 peptides in Drosophila cells, with the highest affinity being for H4K20me1/2 (Scharf et al., 2009). The ATPase activity of valosin-containing protein promotes the release of L3MBTL1 from chromatin; L3MBTL1 binds to H4K20me2, thereby facilitating 53BP1 recruitment (Acs et al., 2011). Other histone modifications can occupy 53BP1 to disrupt its binding with H4K20me2 and thus direct DNA repair. H3K79me2 is primarily responsible for 53BP1 binding in budding yeast (Chen & Zhu, 2016). In human cells, although both methylations H3K79 and H4K20 are able to bind to 53BP1, H4K20 methylation binds more tightly. Additionally, TIP60-catalyzed H4K16ac impacts the ability of 53BP1 to bind neighboring H4K20me2 (Panier & Boulton, 2014; Tang et al., 2013). Similarly, 53BP1 recognizes mononucleosomes containing both H4K20me2 and H2AK15ub as a dimer using its methyl-lysine-binding Tudor domain and UDR motif, respectively (Fradet-Turcotte et al., 2013). Interestingly, the ubiquitylation of H2AK15 by RNF168 can be blocked by Tat-interactive protein 60-kDa (Tip60)-catalyzed acetylation of H2AK15 in cis form (Jacquet et al., 2016); therefore, TIP60 regulates 53BP1-dependent repair through the competitive bivalent modification of chromatin.
H4K20me3 is enriched in constitutive heterochromatin and regulates the stability of telomere and heterochromatin structures (Jorgensen et al., 2013). Because the specific partner proteins of H4K20me3 are unknown, the detailed mechanisms of how H4K20me3 regulates heterochromatin and DNA damage are not understood.
Histone H4 lysine20 demethylation
KDM7B is the first reported demethylase of H4K20me1 that is also linked to the enzymes related to cell cycle progression and proliferation (Liu et al., 2010) (Yatim et al., 2012). The stabilization of histone demethylase KDM7B by ubiquitin specific protease 7 is augmented during DNA damage (Wang et al., 2016). Replication stress results in KDM7B phosphorylation and dissociation from topoisomerase IIβ-binding protein 1 (TOPBP1). Consequently, hypomethylated TOPBP1 facilitates RADiation-sensitive 9-binding to chromatin to fully activate ATR and thus safeguard the genome and protect cells against replication stress (Feng et al., 2020; Ma et al., 2021). Although many studies have observed the role of KDM7B in transcription silencing, further studies are needed to elaborate on the role of KDM7B in the DDR.
Perspectives and concluding remarks
In contrast to DNA methylation, which only exists in higher eukaryotes, histone methylation is conserved in organisms such as Drosophila melanogaster and C. elegans, in which DNA methylation is absent. In addition, histone methylation is a reversible event that is modified by methyltransferases and demethylases. Therefore, dynamic histone methylation is indispensable for maintaining normal biological function. During the past two decades, there has been several progress are reported to be regulated by H4K20 methylation. On one hand, H4K20 methylation is able to directly affect chromatin compaction. On the other hand, different degrees of H4K20me serve as a binding platform for a variety of effectors, and therefore, the modification is associated with diverse functions. The identification of novel proteins that either regulate H4K20 modulators or recognize different degrees of H4K20 methylation will enrich our knowledge of the roles of H4K20 methylation.
Methylation can occur at multiple different sites on the same histone, and one site can incur different modifications. Different combinations of histone modifications can alter the recognition and binding of DDR factors. For example, H4K16ac adjacent to H4K20me2 affects 53BP1 binding through the disruption of H4K16 and Tudor domain interactions (Lu et al., 2019). Because histone methylation marks do not appear in isolation, it is important to determine the collaborative and antagonistic relationships among different histone marks.
The importance of histone methylation is also being demonstrated by emerging evidence that links histone methylation to disease and ageing. At present, a select group of demethylase inhibitors is being evaluated in clinical trials (Wen et al., 2019). One of the most studied inhibitors is tranylcypromine, which efficiently inhibits KDM1A by forming a covalent adduct with the FAD cofactor, and two clinical trials using tranylcypromine to cure acute myeloid leukemia and myelodysplastic syndrome are underway (Jambhekar et al., 2017). The removal of H4K20 methyltransferases results in lethality, and loss of H4K20me3 is a hallmark of many human cancers. However, there are still no specific inhibitors of H4K20me demethylases. Developing highly efficient inhibitors is of significance to transfer our understanding of methylation from the bench to the bedside.
Availability of data and materials
Not applicable.
References
Abbas, T., Shibata, E., Park, J., Jha, S., Karnani, N., & Dutta, A. (2010). CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Molecular Cell, 40(1), 9–21. https://doi.org/10.1016/j.molcel.2010.09.014
Acs, K., Luijsterburg, M. S., Ackermann, L., Salomons, F. A., Hoppe, T., & Dantuma, N. P. (2011). The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature Structural & Molecular Biology, 18(12), 1345–1350. https://doi.org/10.1038/nsmb.2188
Alagoz, M., Katsuki, Y., Ogiwara, H., Ogi, T., Shibata, A., Kakarougkas, A., & Jeggo, P. (2015). SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Research, 43(16), 7931–7944. https://doi.org/10.1093/nar/gkv722
Alonso-de Vega, I., Paz-Cabrera, M. C., Rother, M. B., Wiegant, W. W., Checa-Rodriguez, C., Hernandez-Fernaud, J. R., & Smits, V. A. J. (2020). PHF2 regulates homology-directed DNA repair by controlling the resection of DNA double strand breaks. Nucleic Acids Research, 48(9), 4915–4927. https://doi.org/10.1093/nar/gkaa196
Ayrapetov, M. K., Gursoy-Yuzugullu, O., Xu, C., Xu, Y., & Price, B. D. (2014). DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proceedings of the National Academy of Sciences U S A, 111(25), 9169–9174. https://doi.org/10.1073/pnas.1403565111
Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., & Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410(6824), 120–124. https://doi.org/10.1038/35065138
Beck, D. B., Oda, H., Shen, S. S., & Reinberg, D. (2012). PR-Set7 and H4K20me1: At the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes & Development, 26(4), 325–337. https://doi.org/10.1101/gad.177444.111
Biggar, K. K., & Li, S. S. (2015). Non-histone protein methylation as a regulator of cellular signalling and function. Nature Reviews Molecular Cell Biology, 16(1), 5–17. https://doi.org/10.1038/nrm3915
Black, J. C., Van Rechem, C., & Whetstine, J. R. (2012). Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular Cell, 48(4), 491–507. https://doi.org/10.1016/j.molcel.2012.11.006
Botuyan, M. V., Lee, J., Ward, I. M., Kim, J. E., Thompson, J. R., Chen, J., & Mer, G. (2006). Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell, 127(7), 1361–1373. https://doi.org/10.1016/j.cell.2006.10.043
Brejc, K., Bian, Q., Uzawa, S., Wheeler, B. S., Anderson, E. C., King, D. S., & Meyer, B. J. (2017). Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell, 171(1), 85-1023 e123. https://doi.org/10.1016/j.cell.2017.07.041
Bromberg, K. D., Mitchell, T. R., Upadhyay, A. K., Jakob, C. G., Jhala, M. A., Comess, K. M., & Pappano, W. N. (2017). The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nature Chemical Biology, 13(3), 317–324. https://doi.org/10.1038/nchembio.2282
Cao, L. L., Shen, C., & Zhu, W. G. (2016). Histone modifications in DNA damage response. Science China Life Sciences, 59(3), 257–270. https://doi.org/10.1007/s11427-016-5011-z
Cao, L. L., Wei, F., Du, Y., Song, B., Wang, D., Shen, C., & Zhu, W. G. (2016). ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene, 35(3), 301–313. https://doi.org/10.1038/onc.2015.81
Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., & Zhang, Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science, 298(5595), 1039–1043. https://doi.org/10.1126/science.1076997
Cao, X., Chen, Y., Wu, B., Wang, X., Xue, H., Yu, L., & Chen, C. D. (2020). Histone H4K20 Demethylation by Two hHR23 Proteins. Cell Reports, 30(12), 4152-4164e4156. https://doi.org/10.1016/j.celrep.2020.03.001
Ceccaldi, R., Rondinelli, B., & D’Andrea, A. D. (2016). Repair pathway choices and consequences at the double-strand break. Trends in Cell Biology, 26(1), 52–64. https://doi.org/10.1016/j.tcb.2015.07.009
Centore, R. C., Havens, C. G., Manning, A. L., Li, J. M., Flynn, R. L., Tse, A., & Zou, L. (2010). CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Molecular Cell, 40(1), 22–33. https://doi.org/10.1016/j.molcel.2010.09.015
Chang, S., Yim, S., & Park, H. (2019). The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: Crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism. Experimental & Molecular Medicine, 51(6), 1–17. https://doi.org/10.1038/s12276-019-0230-6
Chen, Y., & Zhu, W. G. (2016). Biological function and regulation of histone and non-histone lysine methylation in response to DNA damage. Acta Biochimica Et Biophysica Sinica (shanghai), 48(7), 603–616. https://doi.org/10.1093/abbs/gmw050
Clauson, C., Scharer, O. D., & Niedernhofer, L. (2013). Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspectives in Biology, 5(10), a012732. https://doi.org/10.1101/cshperspect.a012732
Dimitrova, E., Turberfield, A. H., & Klose, R. J. (2015). Histone demethylases in chromatin biology and beyond. EMBO Reports, 16(12), 1620–1639. https://doi.org/10.15252/embr.201541113
Dulev, S., Tkach, J., Lin, S., & Batada, N. N. (2014). SET8 methyltransferase activity during the DNA double-strand break response is required for recruitment of 53BP1. EMBO Reports, 15(11), 1163–1174. https://doi.org/10.15252/embr.201439434
Edwards, C. R., Dang, W., & Berger, S. L. (2011). Histone H4 lysine 20 of Saccharomyces cerevisiae is monomethylated and functions in subtelomeric silencing. Biochemistry, 50(48), 10473–10483. https://doi.org/10.1021/bi201120q
Fang, J., Feng, Q., Ketel, C. S., Wang, H., Cao, R., Xia, L., & Zhang, Y. (2002). Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Current Biology, 12(13), 1086–1099. https://doi.org/10.1016/s0960-9822(02)00924-7
Fang, R., Barbera, A. J., Xu, Y., Rutenberg, M., Leonor, T., Bi, Q., & Shi, Y. G. (2010). Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Molecular Cell, 39(2), 222–233. https://doi.org/10.1016/j.molcel.2010.07.008
Farooq, Z., Banday, S., Pandita, T. K., & Altaf, M. (2016). The many faces of histone H3K79 methylation. Mutation Research, Reviews in Mutation Research, 768, 46–52. https://doi.org/10.1016/j.mrrev.2016.03.005
Faucher, D., & Wellinger, R. J. (2010). Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genetics. https://doi.org/10.1371/journal.pgen.1001082
Feng, H., Lu, J., Song, X., Thongkum, A., Zhang, F., Lou, L., & Gong, Z. (2020). CK2 kinase-mediated PHF8 phosphorylation controls TopBP1 stability to regulate DNA replication. Nucleic Acids Research, 48(19), 10940–10952. https://doi.org/10.1093/nar/gkaa756
FitzGerald, J. E., Grenon, M., & Lowndes, N. F. (2009). 53BP1: function and mechanisms of focal recruitment. Biochemical Society Transactions, 37(Pt 4), 897–904. https://doi.org/10.1042/BST0370897
Fnu, S., Williamson, E. A., De Haro, L. P., Brenneman, M., Wray, J., Shaheen, M., & Hromas, R. (2011). Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proceedings of the National Academy of Sciences U S A, 108(2), 540–545. https://doi.org/10.1073/pnas.1013571108
Fodor, B. D., Shukeir, N., Reuter, G., & Jenuwein, T. (2010). Mammalian Su(var) genes in chromatin control. Annual Review of Cell and Developmental Biology, 26, 471–501. https://doi.org/10.1146/annurev.cellbio.042308.113225
Fradet-Turcotte, A., Canny, M. D., Escribano-Diaz, C., Orthwein, A., Leung, C. C., Huang, H., & Durocher, D. (2013). 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature, 499(7456), 50–54. https://doi.org/10.1038/nature12318
Gong, F., Clouaire, T., Aguirrebengoa, M., Legube, G., & Miller, K. M. (2017). Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. Journal of Cell Biology, 216(7), 1959–1974. https://doi.org/10.1083/jcb.201611135
Green, E. M., Mas, G., Young, N. L., Garcia, B. A., & Gozani, O. (2012). Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nature Structural & Molecular Biology, 19(3), 361–363. https://doi.org/10.1038/nsmb.2252
Greer, E. L., & Shi, Y. (2012). Histone methylation: A dynamic mark in health, disease and inheritance. Nature Reviews Genetics, 13(5), 343–357. https://doi.org/10.1038/nrg3173
Gurvich, N., Perna, F., Farina, A., Voza, F., Menendez, S., Hurwitz, J., & Nimer, S. D. (2010). L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability. Proceedings of the National Academy of Sciences U S A, 107(52), 22552–22557. https://doi.org/10.1073/pnas.1017092108
Husmann, D., & Gozani, O. (2019). Histone lysine methyltransferases in biology and disease. Nature Structural & Molecular Biology, 26(10), 880–889. https://doi.org/10.1038/s41594-019-0298-7
Huyen, Y., Zgheib, O., Ditullio, R. A., Jr., Gorgoulis, V. G., Zacharatos, P., Petty, T. J., & Halazonetis, T. D. (2004). Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature, 432(7015), 406–411. https://doi.org/10.1038/nature03114
Jackson, S. P., & Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature, 461(7267), 1071–1078. https://doi.org/10.1038/nature08467
Jacobs, S. A., & Khorasanizadeh, S. (2002). Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science, 295(5562), 2080–2083. https://doi.org/10.1126/science.1069473
Jacquet, K., Fradet-Turcotte, A., Avvakumov, N., Lambert, J. P., Roques, C., Pandita, R. K., & Cote, J. (2016). The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Molecular Cell, 62(3), 409–421. https://doi.org/10.1016/j.molcel.2016.03.031
Jambhekar, A., Anastas, J. N., & Shi, Y. (2017). Histone lysine demethylase inhibitors. Cold Spring Harbor Perspectives in Medicine. https://doi.org/10.1101/cshperspect.a026484
Jorgensen, S., Elvers, I., Trelle, M. B., Menzel, T., Eskildsen, M., Jensen, O. N., & Sorensen, C. S. (2007). The histone methyltransferase SET8 is required for S-phase progression. Journal of Cell Biology, 179(7), 1337–1345. https://doi.org/10.1083/jcb.200706150
Jorgensen, S., Schotta, G., & Sorensen, C. S. (2013). Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Research, 41(5), 2797–2806. https://doi.org/10.1093/nar/gkt012
Kang, J. Y., Park, J. W., Hahm, J. Y., Jung, H., & Seo, S. B. (2020). Histone H3K79 demethylation by KDM2B facilitates proper DNA replication through PCNA dissociation from chromatin. Cell Proliferation, 53(11), e12920. https://doi.org/10.1111/cpr.12920
Kapoor-Vazirani, P., Kagey, J. D., & Vertino, P. M. (2011). SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Molecular and Cellular Biology, 31(8), 1594–1609. https://doi.org/10.1128/MCB.00524-10
Karakashev, S., Fukumoto, T., Zhao, B., Lin, J., Wu, S., Fatkhutdinov, N., & Zhang, R. (2020). EZH2 inhibition sensitizes CARM1-high, homologous recombination proficient ovarian cancers to PARP inhibition. Cancer Cell, 37(2), 157-165e156. https://doi.org/10.1016/j.ccell.2019.12.015
Kennedy, R. D., & D’Andrea, A. D. (2006). DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. Journal of Clinical Oncology, 24(23), 3799–3808. https://doi.org/10.1200/JCO.2005.05.4171
Khoury-Haddad, H., Guttmann-Raviv, N., Ipenberg, I., Huggins, D., Jeyasekharan, A. D., & Ayoub, N. (2014). PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proceedings of the National Academy of Sciences U S A, 111(7), E728-737. https://doi.org/10.1073/pnas.1317585111
Khurana, S., Kruhlak, M. J., Kim, J., Tran, A. D., Liu, J., Nyswaner, K., & Oberdoerffer, P. (2014). A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Reports, 8(4), 1049–1062. https://doi.org/10.1016/j.celrep.2014.07.024
Klose, R. J., Kallin, E. M., & Zhang, Y. (2006). JmjC-domain-containing proteins and histone demethylation. Nature Reviews Genetics, 7(9), 715–727. https://doi.org/10.1038/nrg1945
Kramara, J., Osia, B., & Malkova, A. (2018). Break-induced replication: the where, the why, and the how. Trends in Genetics, 34(7), 518–531. https://doi.org/10.1016/j.tig.2018.04.002
Kuser-Abali, G., Gong, L., Yan, J., Liu, Q., Zeng, W., Williamson, A., & Yuan, Z. M. (2018). An EZH2-mediated epigenetic mechanism behind p53-dependent tissue sensitivity to DNA damage. Proceedings of the National Academy of Sciences U S A, 115(13), 3452–3457. https://doi.org/10.1073/pnas.1719532115
Lee, D. H., Kim, G. W., Jeon, Y. H., Yoo, J., Lee, S. W., & Kwon, S. H. (2020). Advances in histone demethylase KDM4 as cancer therapeutic targets. The FASEB Journal, 34(3), 3461–3484. https://doi.org/10.1096/fj.201902584R
Lee, S. H., Oshige, M., Durant, S. T., Rasila, K. K., Williamson, E. A., Ramsey, H., & Hromas, R. (2005). The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proceedings of the National Academy of Sciences U S A, 102(50), 18075–18080. https://doi.org/10.1073/pnas.0503676102
Li, G., Tian, Y., & Zhu, W. G. (2020). The roles of histone deacetylases and their inhibitors in cancer therapy. Frontiers in Cell and Developmental Biology, 8, 576946. https://doi.org/10.3389/fcell.2020.576946
Li, X., Liu, L., Yang, S., Song, N., Zhou, X., Gao, J., & Shi, L. (2014). Histone demethylase KDM5B is a key regulator of genome stability. Proceedings of the National Academy of Sciences U S A, 111(19), 7096–7101. https://doi.org/10.1073/pnas.1324036111
Li, Y., Armstrong, R. L., Duronio, R. J., & MacAlpine, D. M. (2016). Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila. Nucleic Acids Research, 44(15), 7204–7218. https://doi.org/10.1093/nar/gkw333
Li, Z., & Zhu, W. G. (2014). Targeting histone deacetylases for cancer therapy: from molecular mechanisms to clinical implications. International Journal of Biological Sciences, 10(7), 757–770. https://doi.org/10.7150/ijbs.9067
Liu, W., Tanasa, B., Tyurina, O. V., Zhou, T. Y., Gassmann, R., Liu, W. T., & Rosenfeld, M. G. (2010). PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature, 466(7305), 508–512. https://doi.org/10.1038/nature09272
Liu, X., Wang, D., Zhao, Y., Tu, B., Zheng, Z., Wang, L., & Zhu, W. G. (2011). Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc Natl Acad Sci U S A, 108(5), 1925–1930. https://doi.org/10.1073/pnas.1019619108
Long, H., Zhang, L., Lv, M., Wen, Z., Zhang, W., Chen, X., & Li, G. (2020). H2A.Z facilitates licensing and activation of early replication origins. Nature, 577(7791), 576–581. https://doi.org/10.1038/s41586-019-1877-9
Lord, C. J., & Ashworth, A. (2012). The DNA damage response and cancer therapy. Nature, 481(7381), 287–294. https://doi.org/10.1038/nature10760
Lu, X., Tang, M., Zhu, Q., Yang, Q., Li, Z., Bao, Y., & Zhu, W. G. (2019). GLP-catalyzed H4K16me1 promotes 53BP1 recruitment to permit DNA damage repair and cell survival. Nucleic Acids Research, 47(21), 10977–10993. https://doi.org/10.1093/nar/gkz897
Lu, X., Xu, M., Zhu, Q., Zhang, J., Liu, G., Bao, Y., & Zhu, W. G. (2021). RNF8-ubiquitinated KMT5A is required for RNF168-induced H2A ubiquitination in response to DNA damage. The FASEB Journal, 35(4), e21326. https://doi.org/10.1096/fj.202002234R
Ma, S., Cao, C., Che, S., Wang, Y., Su, D., Liu, S., & Shi, L. (2021). PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability. Science Advances. https://doi.org/10.1126/sciadv.abf7684
Mallette, F. A., Mattiroli, F., Cui, G., Young, L. C., Hendzel, M. J., Mer, G., & Richard, S. (2012). RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO Journal, 31(8), 1865–1878. https://doi.org/10.1038/emboj.2012.47
Margueron, R., & Reinberg, D. (2011). The Polycomb complex PRC2 and its mark in life. Nature, 469(7330), 343–349. https://doi.org/10.1038/nature09784
Mehta, R. L., Burdmann, E. A., Cerda, J., Feehally, J., Finkelstein, F., Garcia-Garcia, G., & Remuzzi, G. (2016). Recognition and management of acute kidney injury in the International Society of Nephrology 0by25 Global Snapshot: A multinational cross-sectional study. Lancet, 387(10032), 2017–2025. https://doi.org/10.1016/S0140-6736(16)30240-9
Mosammaparast, N., Kim, H., Laurent, B., Zhao, Y., Lim, H. J., Majid, M. C., & Shi, Y. (2013). The histone demethylase LSD1/KDM1A promotes the DNA damage response. Journal of Cell Biology, 203(3), 457–470. https://doi.org/10.1083/jcb.201302092
Murray, K. (1964). The occurrence of epsilon-N-methyl lysine in histones. Biochemistry, 3, 10–15. https://doi.org/10.1021/bi00889a003
Nishioka, K., Rice, J. C., Sarma, K., Erdjument-Bromage, H., Werner, J., Wang, Y., & Reinberg, D. (2002). PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Molecular Cell, 9(6), 1201–1213. https://doi.org/10.1016/s1097-2765(02)00548-8
Oda, H., Hubner, M. R., Beck, D. B., Vermeulen, M., Hurwitz, J., Spector, D. L., & Reinberg, D. (2010). Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Molecular Cell, 40(3), 364–376. https://doi.org/10.1016/j.molcel.2010.10.011
Oda, H., Okamoto, I., Murphy, N., Chu, J., Price, S. M., Shen, M. M., & Reinberg, D. (2009). Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Molecular and Cellular Biology, 29(8), 2278–2295. https://doi.org/10.1128/MCB.01768-08
Panier, S., & Boulton, S. J. (2014). Double-strand break repair: 53BP1 comes into focus. Nature Reviews Molecular Cell Biology, 15(1), 7–18. https://doi.org/10.1038/nrm3719
Paulsen, R. D., Soni, D. V., Wollman, R., Hahn, A. T., Yee, M. C., Guan, A., & Cimprich, K. A. (2009). A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Molecular Cell, 35(2), 228–239. https://doi.org/10.1016/j.molcel.2009.06.021
Pei, H., Zhang, L., Luo, K., Qin, Y., Chesi, M., Fei, F., & Lou, Z. (2011). MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature, 470(7332), 124–128. https://doi.org/10.1038/nature09658
Pesavento, J. J., Yang, H., Kelleher, N. L., & Mizzen, C. A. (2008). Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Molecular and Cellular Biology, 28(1), 468–486. https://doi.org/10.1128/MCB.01517-07
Peters, A. H., O’Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., & Jenuwein, T. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107(3), 323–337. https://doi.org/10.1016/s0092-8674(01)00542-6
Pfister, S. X., Ahrabi, S., Zalmas, L. P., Sarkar, S., Aymard, F., Bachrati, C. Z., & Humphrey, T. C. (2014). SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Reports, 7(6), 2006–2018. https://doi.org/10.1016/j.celrep.2014.05.026
Qi, H. H., Sarkissian, M., Hu, G. Q., Wang, Z., Bhattacharjee, A., Gordon, D. B., & Shi, Y. (2010). Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature, 466(7305), 503–507. https://doi.org/10.1038/nature09261
Rea, S., Eisenhaber, F., O’Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., & Jenuwein, T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406(6796), 593–599. https://doi.org/10.1038/35020506
Rolando, M., Sanulli, S., Rusniok, C., Gomez-Valero, L., Bertholet, C., Sahr, T., & Buchrieser, C. (2013). Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host & Microbe, 13(4), 395–405. https://doi.org/10.1016/j.chom.2013.03.004
Sakaguchi, A., & Steward, R. (2007). Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. Journal of Cell Biology, 176(2), 155–162. https://doi.org/10.1083/jcb.200607178
Sanders, S. L., Portoso, M., Mata, J., Bahler, J., Allshire, R. C., & Kouzarides, T. (2004). Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell, 119(5), 603–614. https://doi.org/10.1016/j.cell.2004.11.009
Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., & Kouzarides, T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature, 419(6905), 407–411. https://doi.org/10.1038/nature01080
Scharf, A. N., Meier, K., Seitz, V., Kremmer, E., Brehm, A., & Imhof, A. (2009). Monomethylation of lysine 20 on histone H4 facilitates chromatin maturation. Molecular and Cellular Biology, 29(1), 57–67. https://doi.org/10.1128/MCB.00989-08
Schotta, G., Lachner, M., Sarma, K., Ebert, A., Sengupta, R., Reuter, G., & Jenuwein, T. (2004). A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes & Development, 18(11), 1251–1262. https://doi.org/10.1101/gad.300704
Schotta, G., Sengupta, R., Kubicek, S., Malin, S., Kauer, M., Callen, E., & Jenuwein, T. (2008). A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & Development, 22(15), 2048–2061. https://doi.org/10.1101/gad.476008
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., & Shi, Y. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell, 119(7), 941–953. https://doi.org/10.1016/j.cell.2004.12.012
Shi, Y., Sawada, J., Sui, G., el Affar, B., Whetstine, J. R., Lan, F., & Shi, Y. (2003). Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature, 422(6933), 735–738. https://doi.org/10.1038/nature01550
Shibata, A. (2017). Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutation Research, 803–805, 51–55. https://doi.org/10.1016/j.mrfmmm.2017.07.011
Sone, K., Piao, L., Nakakido, M., Ueda, K., Jenuwein, T., Nakamura, Y., & Hamamoto, R. (2014). Critical role of lysine 134 methylation on histone H2AX for gamma-H2AX production and DNA repair. Nature Communications, 5, 5691. https://doi.org/10.1038/ncomms6691
Southall, S. M., Cronin, N. B., & Wilson, J. R. (2014). A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases. Nucleic Acids Research, 42(1), 661–671. https://doi.org/10.1093/nar/gkt776
Takahashi, A., Imai, Y., Yamakoshi, K., Kuninaka, S., Ohtani, N., Yoshimoto, S., & Hara, E. (2012). DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Molecular Cell, 45(1), 123–131. https://doi.org/10.1016/j.molcel.2011.10.018
Tan, M., Luo, H., Lee, S., Jin, F., Yang, J. S., Montellier, E., & Zhao, Y. (2011). Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell, 146(6), 1016–1028. https://doi.org/10.1016/j.cell.2011.08.008
Tang, J., Cho, N. W., Cui, G., Manion, E. M., Shanbhag, N. M., Botuyan, M. V., & Greenberg, R. A. (2013). Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature Structural & Molecular Biology, 20(3), 317–325. https://doi.org/10.1038/nsmb.2499
Tardat, M., Murr, R., Herceg, Z., Sardet, C., & Julien, E. (2007). PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. Journal of Cell Biology, 179(7), 1413–1426. https://doi.org/10.1083/jcb.200706179
Trojer, P., Li, G., Sims, R. J., 3rd., Vaquero, A., Kalakonda, N., Boccuni, P., & Reinberg, D. (2007). L3MBTL1, a histone-methylation-dependent chromatin lock. Cell, 129(5), 915–928. https://doi.org/10.1016/j.cell.2007.03.048
Trojer, P., Zhang, J., Yonezawa, M., Schmidt, A., Zheng, H., Jenuwein, T., & Reinberg, D. (2009). Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase G9a/KMT1C and the Jumonji domain-containing JMJD2/KDM4 proteins. Journal of Biological Chemistry, 284(13), 8395–8405. https://doi.org/10.1074/jbc.M807818200
Tu, B., Bao, Y., Tang, M., Zhu, Q., Lu, X., Wang, H., & Zhu, W.-G. (2020). TIP60 recruits SUV39H1 to chromatin to maintain heterochromatin genome stability and resist hydrogen peroxide-induced cytotoxicity. Genome Instability & Disease, 1(6), 339–355. https://doi.org/10.1007/s42764-020-00025-8
Tuzon, C. T., Spektor, T., Kong, X., Congdon, L. M., Wu, S., Schotta, G., & Rice, J. C. (2014). Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Reports, 8(2), 430–438. https://doi.org/10.1016/j.celrep.2014.06.013
Uckelmann, M., & Sixma, T. K. (2017). Histone ubiquitination in the DNA damage response. DNA Repair (Amst), 56, 92–101. https://doi.org/10.1016/j.dnarep.2017.06.011
Van, H. T., & Santos, M. A. (2018). Histone modifications and the DNA double-strand break response. Cell Cycle, 17(21–22), 2399–2410. https://doi.org/10.1080/15384101.2018.1542899
Wagner, E. J., & Carpenter, P. B. (2012). Understanding the language of Lys36 methylation at histone H3. Nature Reviews Molecular Cell Biology, 13(2), 115–126. https://doi.org/10.1038/nrm3274
Walport, L. J., Hopkinson, R. J., Chowdhury, R., Zhang, Y., Bonnici, J., Schiller, R., & Schofield, C. J. (2018). Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26. FEBS Letters, 592(19), 3264–3273. https://doi.org/10.1002/1873-3468.13231
Wang, D., Zhou, J., Liu, X., Lu, D., Shen, C., Du, Y., & Zhu, W. G. (2013). Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proceedings of the National Academy of Sciences U S A, 110(14), 5516–5521. https://doi.org/10.1073/pnas.1216596110
Wang, Q., Ma, S., Song, N., Li, X., Liu, L., Yang, S., & Shi, L. (2016). Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. The Journal of Clinical Investigation, 126(6), 2205–2220. https://doi.org/10.1172/JCI85747
Weirich, S., Kudithipudi, S., & Jeltsch, A. (2016). Specificity of the SUV4-20H1 and SUV4-20H2 protein lysine methyltransferases and methylation of novel substrates. Journal of Molecular Biology, 428(11), 2344–2358. https://doi.org/10.1016/j.jmb.2016.04.015
Wen, H., Li, G., Chen, Y., Li, M., Lv, Y., Li, Z., & Zhu, W. G. (2019). A specific assay for JmjC domain-containing lysine demethylase and its application to inhibitor screening. Science China Life Sciences, 62(10), 1404–1408. https://doi.org/10.1007/s11427-019-9582-9
Wu, H., Siarheyeva, A., Zeng, H., Lam, R., Dong, A., Wu, X. H., & Min, J. (2013). Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2. FEBS Letters, 587(23), 3859–3868. https://doi.org/10.1016/j.febslet.2013.10.020
Yang, Q., Zhu, Q., Lu, X., Du, Y., Cao, L., Shen, C., & Zhu, W. G. (2017). G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proceedings of the National Academy of Sciences U S A, 114(30), E6054–E6063. https://doi.org/10.1073/pnas.1700694114
Yatim, A., Benne, C., Sobhian, B., Laurent-Chabalier, S., Deas, O., Judde, J. G., & Benkirane, M. (2012). NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Molecular Cell, 48(3), 445–458. https://doi.org/10.1016/j.molcel.2012.08.022
Zhang, J., Lu, X., MoghaddamKohi, S., Shi, L., Xu, X., & Zhu, W. G. (2021). Histone lysine modifying enzymes and their critical roles in DNA double-strand break repair. DNA Repair (Amst), 107, 103206. https://doi.org/10.1016/j.dnarep.2021.103206
Zhang, Y., Chang, J. F., Sun, J., Chen, L., Yang, X. M., Tang, H. Y., & Sun, F. L. (2018). Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. Journal of Cell Science. https://doi.org/10.1242/jcs.215525
Zhu, Q., Yang, Q., Lu, X., Wang, H., Tong, L., Li, Z., & Zhu, W. G. (2021). SETD2-mediated H3K14 trimethylation promotes ATR activation and stalled replication fork restart in response to DNA replication stress. Proceedings of the National Academy of Sciences USA. https://doi.org/10.1073/pnas.2011278118
Acknowledgements
This work was supported by National Key R&D Program of China [2017YFA0503900]; National Natural Science Foundation of China [32090030, 81720108027, 81530074, 81972697]; Science and Technology Program of Guangdong Province in China [2017B030301016]; Shenzhen Municipal Commission of Science and Technology Innovation [JCYJ20170818092450901, JCYJ20200109114214463]. The authors would like to thank Dr. Jessica Tamanini of ETediting, UK for English language editing prior to submission.
Author information
Sara Moghaddam Kohi and Tingting Feng contributed equally to this work.
Affiliations
Guangdong Key Laboratory of Genome Instability and Human Disease Prevention, Department of Biochemistry and Molecular Biology, School of Medicine, Shenzhen University, Shenzhen, 518055, China
Sara Moghaddam Kohi, Tingting Feng, Yuan Tian & Wei-Guo Zhu
Shenzhen Bay Laboratory, School of Medicine, Shenzhen University, Shenzhen, 518055, China
Yuan Tian & Wei-Guo Zhu
Corresponding authors
Correspondence to Yuan Tian or Wei-Guo Zhu.
Ethics declarations
Conflict of interest
Author Wei-Guo Zhu is Editor-in-Chief of Genome Instability & Disease.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kohi, S.M., Feng, T., Tian, Y. et al. Biological function and regulation of histone 4 lysine 20 methylation in DNA damage response. GENOME INSTAB. DIS. 3, 33–46 (2022). https://doi.org/10.1007/s42764-022-00063-4
Received13 December 2021
Revised26 January 2022
Accepted31 January 2022
Published07 February 2022
Issue DateFebruary 2022
DOIhttps://doi.org/10.1007/s42764-022-00063-4
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable linkKeywords
DNA damage response
Histone modifications
Lysine methylation
H4K20 methylation








用户登录
还没有账号?
立即注册