Ubiquitin carboxyl-terminal hydrolase 11 promotes autophagy by de-ubiquitinating and stabilizing Beclin-1
Original Research Paper
Open Access
Zheng Li, Shaohong Rao, Chunwei Song, Min Zhu, Hongchang Zhao, Shuping Yuan, Bin Peng & Xingzhi Xu
Genome Instability & Disease 3, 47–55 (2022)
Abstract
Autophagy is a major degradation process that degrades and recycles cytoplasmic materials through lysosome for maintaining cellular homeostasis. Dysregulated autophagy is linked with numerous human diseases including cancer. Autophagy marker protein B-cell lymphoma-2 interacting protein 1 (Beclin-1) is essential for autophagosome initiation and maturation. Recently, Ubiquitin carboxyl-terminal hydrolase 11 (USP11) has been reported to promote or inhibit autophagy without identification of any direct target. Here through biochemical reaction in vitro, we demonstrate that USP11 directly interacts with Beclin-1. Both in vitro and in vivo de-ubiquitination assays revealed that USP11 de-ubiquitinates Beclin-1. USP11-mediated de-ubiquitination stabilized Beclin-1 and enhanced the formation of the autophagy-specific class III phosphatidylinositol 3-kinase complexes 1 and 2, thereby promoting autophagy. Together, our results demonstrated that USP11 promotes autophagy under unperturbed conditions by de-ubiquitinating and stabilizing Beclin-1 which may serve as a therapeutic target for autophagy-related diseases.
Introduction
Autophagy is an evolutionarily conserved cellular recycling process that degrades cytoplasmic proteins, macromolecules, and unwanted organelles through lysosomal degradation to maintain metabolic homeostasis (Rabinowitz & White, 2010; Yang & Klionsky, 2020). In response to different stressors, such as nutrient deprivation and growth factor depletion, autophagy is activated, facilitating cell tolerance of the microenvironment, which prevents cell damage and promotes survival (White & DiPaola, 2009). In mammalian cells, there are three types of autophagy, namely, chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy (Yang & Klionsky, 2010). In the CMA pathway, proteins containing a KFERQ-like motif are recognized by the chaperone HSC70 and cochaperons and delivered to the lysosome for degradation (Dong et al., 2021; Kaushik & Cuervo, 2018; Yang et al., 2019). In microautophagy, the lysosomal membrane invaginates long-lived proteins into the lysosome for degradation through membrane fission (Schuck, 2020). Macroautophagy has been studied extensively and differs from CMA and microautophagy. In the macroautophagy pathway, autophagic cargos need to be packaged into double-membrane vesicles and subsequently transported to the lysosome (Dikic & Elazar, 2018). In brief, the macroautophagy pathway comprises five stages: phagophore initiation and nucleation, expansion, maturation, fusion with the lysosome, and degradation.
Multiple protein complexes are involved in macroautophagy (Dikic & Elazar, 2018). The autophagy marker protein B-cell lymphoma-2 interacting protein 1 (Beclin-1), the lipid kinase VPS34, and the scaffold protein VPS15 constitute core components of the autophagy-specific class III phosphatidylinositol 3-kinase complex (PI3KC3) (Galluzzi & Green, 2019). Beclin-1 recruits one of two positive regulators, ATG14 or UVRAG, in a mutually exclusive manner to serve as the fourth subunit for PI3KC3 complex 1 (PI3KC3-C1) or PI3KC3 complex 2 (PI3KC3-C2), respectively. PI3KC3-C1 is involved in the initiation of autophagosomes and PI3KC3-C2 in the maturation of autophagosomes and endocytosis (Baskaran et al., 2014). During this process, Beclin-1 undergoes multiple functional post-translational modifications, including phosphorylation and ubiquitination. Death-associated protein kinase 1 (DAPK1) phosphorylates Beclin-1 at Thr-119 and reduces Beclin-1 interaction with the apoptosis regulator B-cell lymphoma-2 (Bcl-2), thus promoting autophagy (Zalckvar et al., 2009). The “Lys-11”-linked polyubiquitination of Beclin-1 is mediated by NEDD4 and leads to beclin1 degradation (Platta et al., 2012). Expansion of the CAG (poly-glutamine) repeat in the ATXN3 coding region leads to spinocerebellar ataxia, a clinically and genetically heterogeneous group of cerebellar disorders; whereas normal ATXN3 protein de-ubiquitinates and stabilizes Beclin-1, promoting autophagy, while expansion of the poly-glutamine repeat impairs its function (Ashkenazi et al., 2017).
USP11 has been demonstrated to participate in multiple cellular pathways such as cortical neurogenesis (Chiang et al., 2021), R-loop regulation (Jurga et al., 2021), and DNA repair (Ting et al., 2019). In a proteomic screen, USP11 was identified as a protein that interacts with Beclin-1, VPS34, ATG10, ATG12, and ATG16L1, suggesting that USP11 may function in autophagy (Behrends et al., 2010). Recently, USP11 has been reported to activate autophagy by stabilizing valosin-containing protein (VCP) (Sun et al., 2021). In contrast, Qiao et al. demonstrated that USP11 inhibits autophagy by activating the ERK/mTOR pathway (Qiao et al., 2021). While this manuscript was under preparation, Basic et al. reported that, within the hTERT-immortalized retinal pigment epithelial cell line hTERT RPE-1, USP11 interacts with the PI3KC3-C1 complex and reduces its association with nuclear receptor-binding factor 2 (NRBF2), thus decreasing the lipid kinase activity of PI3KC3-C1 (Basic et al., 2021). Furthermore, USP11 interacts with and stabilizes serine/threonine kinase mTOR, thereby compromising autophagy (Basic et al., 2021). All these studies failed to identify the direct target(s) for USP11, and these conflicting results require clarification.
In this study, we aimed to clarify if USP11 promotes or suppresses autophagy by identification of its direct target(s). We found that USP11 promotes autophagy by directly binding with and de-ubiquitinating Beclin-1, which leads to Beclin-1 stabilization and PI3KC3-1/2 complex formation.
Results
USP11 promotes autophagy in a catalytic activity-dependent manner
Given that the studies before failed to demonstrate if restoring USP11 expression and/or the catalytic activity of USP11 can erase the USP11 depletion-associated autophagic phenotypes, we stably expressed a short hairpin RNA (shRNA) construct targeting the coding region of the USP11 gene in HeLa cells. The resulting USP11 knockdown led to an accumulation of autophagy receptor P62, and there was a decrease in the LC3-II/I ratio on immunoblotting analysis (Fig. 1a) and decreased LC3-II puncta formation on immunocytochemical analysis (Fig. 1b). Re-expression of the shRNA-resistant form of wild type HA-tagged USP11 (HA-USP11), but not the catalytically inactive mutant HA-USP11 (C318S), restored the USP11 depletion-induced changes to P62 levels, the LC3-II/I ratio, and LC3-II puncta formation (Figs. 1a and 1b). Conversely, overexpression of wild type HA-USP11, but not the HA-USP11 (C318S) mutant, increased the LC3-II/I ratio while decreasing P62 protein levels (Fig. 1c). Taken together, our results demonstrated that USP11 promotes autophagy in an enzymatic activity-dependent manner.
Fig. 1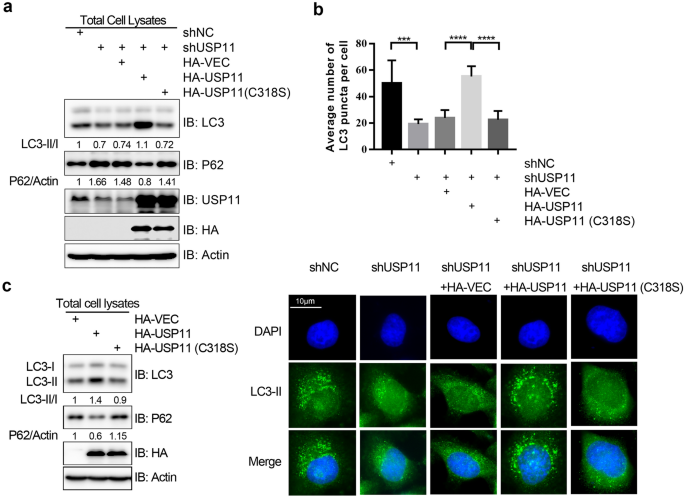
USP11 promotes autophagy in a catalytic activity-dependent manner. a USP11 depletion inhibits autophagy. Western blot showing the expression of HA-tagged wild type and enzymatic activity defective mutant (C318S) USP11, P62, and LC3 in normal control (NC) and vector control (VEC) HeLa cells and HeLa cells treated with short hairpin RNA (shRNA) to USP11 (shUSP11). P62 protein levels were normalized to actin. b USP11 depletion decreases LC3 puncta formation. LC3 puncta were measured by immunofluorescence assay and further quantified by ImageJ software. A two-tailed Student’s t test was used to determine the statistical significance of group differences. *p < 0.05, **p < 0.01, ****p < 0.001. c USP11 overexpression promotes autophagy. Western blot showing the expression of HA-tagged wild type and enzymatic activity defective mutant (C318S) USP11, P62, and LC3 in normal HeLa cells. P62 protein levels were normalized to actin
Full size imageUSP11 directly interacts with Beclin-1 and the interaction is induced by starvation
Three recent studies failed to identify the direct target of USP11-depletion-mediated autophagy (Basic et al., 2021; Qiao et al., 2021; Sun et al., 2021). We sifted through the interaction networks of the human autophagy system described by Behrends et al. and found candidate interactors of USP11, including Beclin-1 and VPS34 (Behrends et al., 2010). Co-immunoprecipitation assays revealed that endogenous USP11 was present in the immunocomplex of endogenous Beclin-1 in HeLa cells (Fig. 2a), and HA-tagged Beclin-1 (HA-Beclin-1) was present in the FLAG-USP11 immunocomplex in HeLa cells co-expressing HA-Beclin-1 and FLAG-USP11 (Fig. 2b). Using GST pulldown assays, we found that bacterially produced GST-USP11 was able to pull down bacterially produced HIS-Beclin-1 (Fig. 2c), indicating that USP11 directly interacts with Beclin-1. Upon glucose starvation treatment, both exogenous and endogenousinteractions between USP11 and Beclin-1 significant increased (Figs. 2d and 2e). We also found a positive correlation (p-Value < 0.05, R-Value > 0.3) between Beclin-1 and USP11 gene expression in 15 types of tumors from the GEPIA database (http://gepia.cancer-pku.cn) (Fig. 2f). Therefore, we concluded that USP11 directly interacts with Beclin-1, and the interaction is induced by starvation.
Fig. 2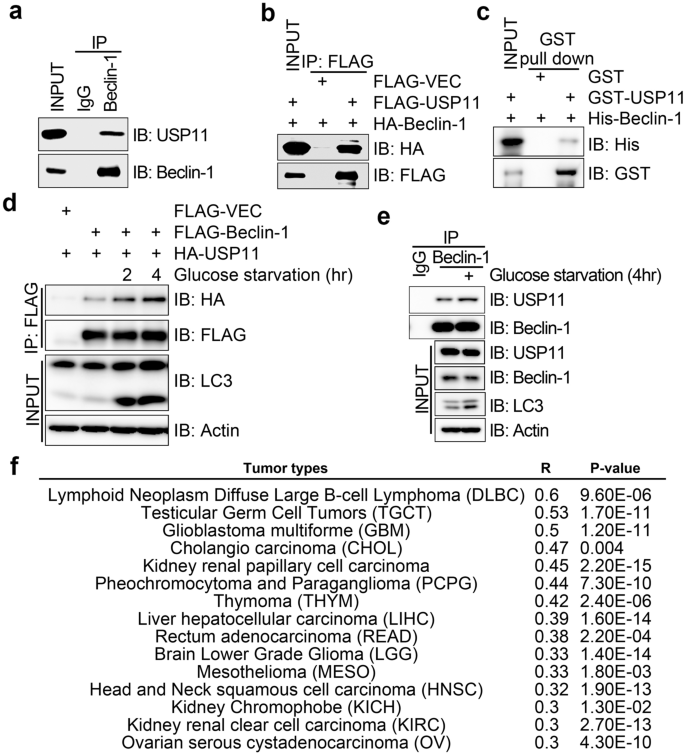
USP11 directly interacts with Beclin-1 and this interaction is induced by starvation. a Endogenous USP11 interacts with Beclin-1. Western blot showing the endogenous USP11 in the anti-Beclin-1 immunoprecipitates but not the anti-IgG (negative control) immunoprecipitates from HEK293T cells. b Ectopic tagged USP11 interacts with Beclin-1. Western blot showing the HA-Beclin-1 in the anti-FLAG immunoprecipitates from HEK293T cells overexpressing HA-Beclin-1 and with or without FLAG-USP11. c USP11 directly interacts with Beclin-1. Western blot showing the bacterial produced His-Beclin-1 in the anti-GST immunoprecipitates that containing GST-USP11 but not the GST (negative control) group. d,e. Starvation stimulates USP11 interaction with Beclin-1. Before harvest, cell culture medium was removed and the cells were washed with PBS for 3 times and then emersed in fresh medium without glucose. Total cell lysates were extracted and subjected to immunoprecipitation with anti-FLAG (d) or anti-Beclin-1 (e) antibody followed by immunoblotting with antibodies as indicated. f Data were collected from GEPIA database (http://gepia.cancer-pku.cn/); 15 types of tumor (33 types of tumor in total) exhibited gene correlation (R-value > 3, p value < 0.05) between Beclin-1 and USP11
Full size imageUSP11 de-ubiquitinates and stabilizes Beclin-1
Given that USP11 directly interacts with Beclin-1 (Fig. 2), we reasoned that Beclin-1 could be a substrate for USP11. To test this hypothesis, we co-expressed FLAG-Beclin-1 and HA-tagged ubiquitin (HA-Ub) along with Myc-USP11 or catalytically inactive mutant Myc-USP11 (C318S) in HEK293T cells. Co-expressing Myc-USP11 greatly reduced the ubiquitination levels of Beclin-1, whereas co-expressing Myc-USP11 (C318S) did not have any obvious impact on the ubiquitination levels of Beclin-1 (Fig. 3a). This enzymatic activity- dependent de-ubiquitination mediated by USP11 was also observed on endogenous Beclin-1 (Fig. 3b). Furthermore, inhibition of USP11 expression by siRNA increased Beclin-1 ubiquitination (Fig. 3c). In vitro de-ubiquitination assays showed that bacterially produced GST-USP11, but not GST-USP11 (C318S), efficiently removed the polyubiquitin chains from FLAG-Beclin-1, which was enriched by immunoprecipitation with an anti-FLAG antibody from HEK293T cells co-expressing FLAG-Beclin-1 and HA-Ub (Fig. 3d). Thus, we concluded that USP11 de-ubiquitinates Beclin-1.
Fig. 3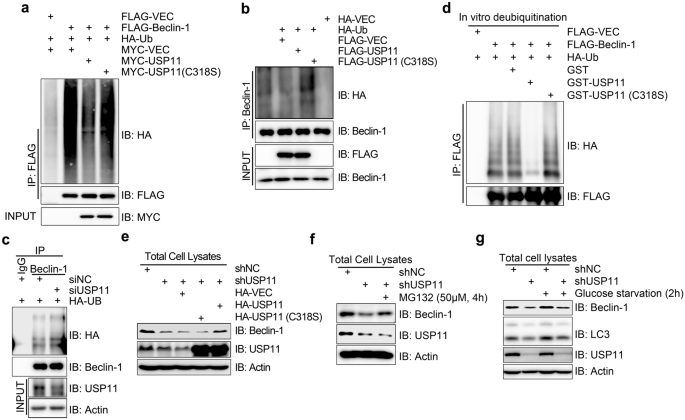
USP11 de-ubiquitinates and stabilizes Beclin-1. a USP11 de-ubiquitinates exogenous Beclin-1 in vivo. Western blot showing the Myc-USP11 but not the Myc-USP11 (C318S) significantly decreased HA signal (ubiquitination of FLAG-Belcin-1) in anti-FLAG immunoprecipitates from HEK293T cells overexpressing HA-ubiquitin and with or without FLAG-Beclin-1. b USP11 de-ubiquitinates endogenous Beclin-1 in vivo. Western blot showing the FLAG-USP11 but not the FLAG-USP11 (C318S) significantly decreased HA signal (ubiquitination of endogenous Belcin-1) in anti-Beclin-1 immunoprecipitates from HEK293T cells overexpressing HA-ubiquitin. c Inhibition of USP11 expression increases Beclin-1 ubiquitination. Western blot showing the inhibition of USP11 expression by siRNA increased HA signal (ubiquitination of endogenous Belcin-1) in anti-Beclin-1 immunoprecipitates from HEK293T cells overexpressing HA-ubiquitin. d USP11 de-ubiquitinates Beclin-1 in vitro. Western blot showing the GST-USP11 but not the GST-USP11 (C318S) significantly decreased HA signal (ubiquitination of Belcin-1) in anti-FLAG immunoprecipitates from HEK293T cells overexpressing HA-ubiquitin and with or without FLAG-Beclin-1. e Wild type USP11, but not the C318S mutant, rescues Beclin-1 protein stability in shUSP11 cells. Western blot showing the expression of Beclin-1 in shUSP11 HeLa cell that reconstituted with HA-tagged vector control, wild type and enzymatic activity defective mutant (C318S) USP11. f MG132 treatment blocks Beclin-1 degradation in USP11-depleted cells. Cells were treated with 50 μM MG132 for 4 h and harvested for western blot detection. g USP11-depletion leads to Beclin-1 degradation. shNC or shUSP11 HeLa cells were treated with or without glucose starvation. Total cell lysates were harvested, followed by immunoblotting with indicated antibodies
Full size imageReportedly, Beclin-1 ubiquitination is a target for proteasomal degradation (Han et al., 2018; Li et al., 2021; Pei et al., 2017). We thus speculated that the USP11-mediated de-ubiquitination of Beclin-1 stabilizes the protein. Indeed, when we inhibited USP11 expression in HeLa cells by shRNA treatment, Beclin-1 protein levels reduced, and this reduction was rescued by the re-expression of wild type HA-USP11, but not the catalytically inactive mutant HA-USP11 (C318S) (Fig. 3e). This USP11-depletion-induced reduction of Beclin-1 protein levels was also restored by pretreating the cells with MG-132—a potent cell-permeable proteasome inhibitor (Fig. 3f). We also found that the USP11-knockdown-induced reduction in Beclin-1 protein levels occurred both under unperturbed and glucose-starvation-induced autophagy conditions (Fig. 3g). Our findings therefore indicate that Beclin-1 is a substrate of USP11, and USP11-mediated de-ubiquitination stabilizes Beclin-1.
USP11 promotes the formation of autophagy-specific PI3KC3 complexes
The scaffolding protein Beclin-1 brings in one of two positive regulators, ATG14 or UVRAG, to serve as the fourth subunit in a mutually exclusive manner to form PI3KC3-C1 and PI3KC3-C2, respectively. We found that the depletion of USP11 expression in HeLa cells decreased Beclin-1 interactions with VPS15, VPS34, ATG14, and UVRAG (Fig. 4a), and this decrease was rescued by the re-expression of HA-USP11, but not that of the catalytically inactive mutant HA-USP11 (C318S) (Fig. 4a). Conversely, overexpressing HA-USP11, but not HA-USP11 (C318S), in HEK293T cells increased the amount of Beclin-1-VPS34-VPS15-ATG14/UVRAG complex formed (Fig. 4b). Taken together, our work has uncovered the direct role of USP11 in autophagy, which involves its de-ubiquitination of Beclin-1 to promote PI3KC3 complex formation.
Fig. 4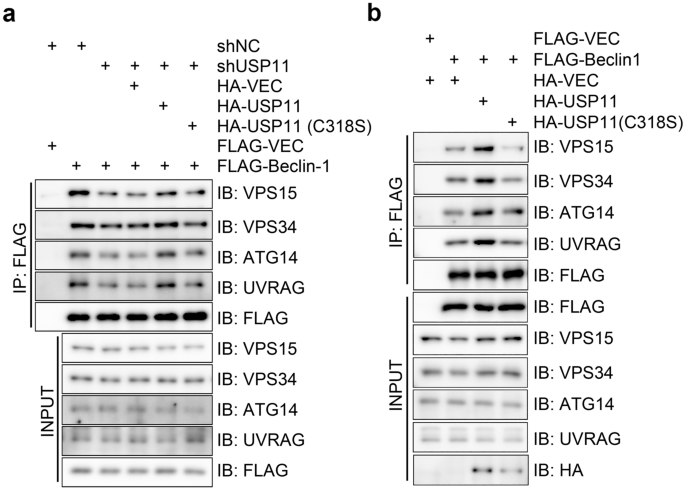
USP11 promotes formation of autophagy-specific PI3KC3 complexes. a USP11-depletion decreases Beclin-1 interaction with other subunits of autophagy-specific class III phosphatidylinositol 3-kinase complexes (PI3KC3). Western blot showing the VPS15, VPS34, ATG14 and UVRAG in the anti-FLAG immunoprecipitates from shUSP11 HeLa cell that reconstituted with HA-tagged vector control, wild type, enzymatic activity defective mutant (C318S) USP11 and with or without FLAG-Beclin-1. b USP11-overexpression promotes Beclin-1 interaction with other subunits of PI3KC3. Western blot showing the VPS15, VPS34, ATG14 and UVRAG in the anti-FLAG immunoprecipitates from HEK293T cells that overexpressing HA-tagged vector control, wild type, enzymatic activity defective mutant (C318S) USP11 and with or without FLAG-Beclin-1
Full size imageDiscussion
It was reported that, without rescue experiments, the inhibition of USP10 or USP13 expression by three independent siRNA oligos destabilized Beclin-1, whereas overexpressing USP10 or USP13, but not the catalytically inactive mutant, reduced Beclin-1 ubiquitination in vivo (Liu et al., 2011). While this manuscript was under preparation, another group also found evidence that USP11 de-ubiquitinates and stabilizes Beclin-1 to promote autophagy and subsequent autophagy-induced ferroptosis (Rong et al., 2021). These studies, along with previously mentioned reports of autophagy modulation by USP11, failed to provide in vitro evidence of a direct interaction between USP11 and Beclin-1 or positive in vitro de-ubiquitination assays of USP11/Beclin-1 (Basic et al., 2021; Qiao et al., 2021; Sun et al., 2021). Thus, these studies could not exclude the possibility that USP11-mediated autophagy is executed through its interacting de-ubiquitinases such as USP4 (L. Zhang et al., 2012), USP7 (Georges et al., 2018), USP15 (Sowa et al., 2009), and OTUD5 (Sowa et al., 2009). In this study, we have provided strong evidence to support the hypothesis that USP11 promotes autophagy by directly de-ubiquitinating and stabilizing Beclin-1. These lines of evidence include results that show (1) USP11 directly interacts with Beclin-1; (2) USP11 de-ubiquitinates Beclin-1 in vitro; and (3) the USP11-mediated de-ubiquitination/stabilization of Beclin-1 and formation of the pro-autophagy PI3KC3 complexes are rescued by the re-expression of wild type USP11, but not the catalytically inactive mutant.
Upregulated USP11 expression has been reported in patient cohorts of gastric cancer (Liu et al., 2021), breast cancer (Bayraktar et al., 2013; Zhou et al., 2017), estrogen receptor alpha-positive breast cancer (Dwane et al., 2020), ovarian cancer (Wang et al., 2019), hepatocellular carcinoma (Zhang et al., 2018), and colorectal cancer (Huang et al., 2021). In addition, high USP11 expression correlated with shorter survival times in patient cohorts of breast cancer (Bayraktar et al., 2013; Garcia et al., 2018; Zhou et al., 2017), estrogen receptor alpha-positive breast cancer (Dwane et al., 2020), ovarian cancer (Wang et al., 2019), hepatocellular carcinoma (Zhang et al., 2018), and colorectal cancer (Huang et al., 2021). We also investigated the relationship between USP11 and Beclin-1 gene expression in 33 types of TCGA tumors from GEPIA database, and there was a positive correlation between the genes in 15 of the tumor types, each exhibiting R values greater than 3. These results suggest that USP11 may functionally correlate with Beclin-1 in tumor cells.
Several studies have demonstrated that autophagy acts to promote tumor survival and growth in advanced cancers (Dikic & Elazar, 2018), and after exploring the GEPIA database (http://gepia.cancer-pku.cn/), we discovered that high Beclin-1 expression correlates with shorter survival times in patient cohorts of ovarian cancer, kidney chromophobe, and lower grade brain glioma (Fig. S1). This suggests that autophagy may be linked to the poor prognosis following some tumor therapeutics. Moreover, the USP11-mediated stabilization of VCP activated autophagy and conferred colorectal cancer resistance to fluorouracil treatment (Sun et al., 2021). Taken together, we demonstrate that Beclin-1 is a direct target for USP11 and USP11-mediated de-ubiquitination and stabilization of Beclin-1 leads to assembling of PI3KC3-C1 and PI3KC3-C2 complex, ultimately promoting autophagy. Therefore, targeting USP11 for inhibition could serve as a therapeutic strategy for autophagy-related diseases.
Materials and methods
Cell culture
Human HEK293T and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin in an atmosphere containing 5% CO2 at 37 ℃.
Antibodies
The following antibodies were used in this study: Anti-FLAG M2(F3165, Sigma), Anti-Beclin-1 (A302-566A/567A, Bethyl), Anti-HA Tag (AE036, ABclonal), Anti-VPS15 (A302-571A, Bethyl), Anti-VPS34 (A12295, ABclonal), Anti-UVRAG (A8462, ABclonal), Anti-LC3 (A19665, ABclonal), Anti-P62 (A302-856A, Bethyl), Anti-Actin (AC026, ABclonal), Anti-His (D2913, MBL), Anti-GST (M2093, MBL), Anti-USP11 (A301-613A), Anti-ATG14 (A7526, ABclonal).
Glucose starvation
For starvation induction, glucose-free medium (11,966–025, Gibco) were used. Before harvested, cultured cells were quickly washed with PBS for 3 times and added with glucose-free medium for indicated time.
Plasmids and transfection
A full-length cDNA clone encoding human Beclin-1 was amplified by PCR and subcloned into the pcDNA3.0 3XFLAG vector and pcDNA3.0 3xHA vector. Wild type USP11 cDNA was cloned into the pcDNA3.0-HA vector, pcDNA3.0-FLAG vector and pcDNA3.1-Myc vector. The cDNA of Ubiquitin was cloned into the pcDNA3.1-Myc vector. Point mutations in the catalytically inactive USP11 (C318S) gene were generated using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme, Nanjing, China. The siRNA oligonucleotide against USP11 (si-USP11 sequence: CUGAAAUGAAAAAAAGGUA) were purchased from Guangzhou RiboBio. The shRNA sequence targeting USP11 was 5′-TCGCGGTTTCCAACCATTATG-3′. Bacteria expressing His-tagged Beclin-1 and GST-tagged USP11 were generated using the pET28a (Invitrogen) and pGEX-4T-1 (GE Healthcare) systems. Plasmid transfections were performed with polyethyleneimine (PEI).
Undenatured co-immunoprecipitation
Cells were harvested and lysed with lysis buffer (150 mM NaCl; 50 mM Tris, pH 7.5; 0.5% NP-40; 5 mM EDTA) at 4 ℃ for 30 min. Further the lysates were centrifuge with 13000g to collect the supernatant for immunoprecipitation with antibody at 4 °C overnight. Then, protein A/G magnetic beads were added to the cell lysates and incubated at 4 °C for 1 h. The beads were washed with cold lysis buffer three times, for 5 min each time, on a rotating mixer.
Denatured co-immunoprecipitation
Immunoprecipitation assays in denaturing conditions (SDS-IP) were executed as previously described (Yoo et al., 2014). Briefly, the harvested cells were lysed with lysis buffer (150 mM Tris, pH 8.0, 5% SDS, 30% glycerol) at 100℃ for 10 min before being digested with benzonuclease (Sigma) at room temperature for 30 min. After centrifugation (13000g, 10 min), the supernatant was diluted with Buffer A (50 mM Tris–HCl [pH 8], 150 mM NaCl, 1% Triton X-100, 1 × protease inhibitor cocktail, and 2 mM NEM) to 20 times dilution and immunoprecipitated with appropriate antibodies.
De-ubiquitination in vitro
Cells transfected with FLAG-Beclin-1 and HA-Ub were harvested 48 h after transfection and lysed with 400 mM NaCl washing buffer (400 mM NaCl; 50 mM Tris, pH 7.5; 0.5% NP-40; 5 mM EDTA). Flag-Beclin-1 protein and its modified forms were pulled down with FLAG-M2 beads. After washing with 400 mM washing buffer three times, the beads were incubated with GST/GST-USP11/GST-USP11 (C318S) in de-ubiquitination buffer (50 mM Tris–HCl [pH 7.4], 50 mM MgCl2, 1 mM DTT) at 37 ℃ for 1 h. After incubation, the beads were washed with 400 mM washing buffer three times.
Immunofluorescence
HeLa cells were washed with PBS three times and fixed with 4% paraformaldehyde at room temperature for 10 min. Then the cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min. After permeabilization, the cells were blocked with 2% BSA in PBST at room temperature for 30 min and further incubated with the primary antibody overnight followed by the secondary antibody for 1 h. After washing with PBST three times, DAPI was added, and the coverslips (CITOGLAS) were covered with antifade. And images were captured with OLYMPUS fluorescence microscope (CKX53).
Western blot
Total protein from cells was extracted with 150 mM NaCl lysis buffer as described earlier. Samples were separated using SDS-PAGE and transferred to PVDF membranes. After skim milk blocking, the membranes were incubated with primary antibody at 4 °C overnight. The membranes were incubated with secondary antibodies for 1 h at room temperature, then visualized using electro chemiluminescence detection reagents (SuperSignal™ West Pico PLUS, Thermo, lot#34,580).
Gene correlation
Gene correlation between USP11 and Beclin-1 in tumors was analyzed online by GEPIA (http://gepia.cancer-pku.cn). This website performs pair-wise gene expression correlation analysis for given sets of TCGA expression data, using Pearson method. Cases that p-Value < 0.05, R-Value > 0.3 were selected as convincible correlation between USP11 and Beclin-1.
Kaplan–Meier survival analysis
The survival analysis is done online (http://gepia.cancer-pku.cn) to analyze correlations between levels of Beclin-1 with overall survival (OS) in cancers. All survival diagrams shown in the manuscript were analyzed by GEPIA using Log-rank test, a.k.a the Mantel–Cox test.
Statistical analysis
Data were analyzed by Prism 7.0 software (GraphPad Prism). A two-tailed Student’s t test was used to determine the statistical significance. All data are presented as the means ± s.d. or means ± s.e.m. as indicated, and a p value of < 0.05 was considered statistically significant.
Data availability
All data generated or analyzed during this study are included in this manuscript. Materials described in this manuscript are available upon request.
Code availability
Not applicable.
References
Ashkenazi, A., Bento, C. F., Ricketts, T., Vicinanza, M., Siddiqi, F., Pavel, M., Squitieri, F., Hardenberg, M. C., Imarisio, S., Menzies, F. M., & Rubinsztein, D. C. (2017). Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature, 545(7652), 108–111. https://doi.org/10.1038/nature22078
Basic, M., Hertel, A., Bajdzienko, J., Bonn, F., Tellechea, M., Stolz, A., Kern, A., Behl, C., & Bremm, A. (2021). The deubiquitinase USP11 is a versatile and conserved regulator of autophagy. Journal of Biological Chemistry, 297(5), 101263. https://doi.org/10.1016/j.jbc.2021.101263
Baskaran, S., Carlson, L. A., Stjepanovic, G., Young, L. N., Kim, D. J., Grob, P., Stanley, R. E., Nogales, E., & Hurley, J. H. (2014). Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife, 3. doi:https://doi.org/10.7554/eLife.05115
Bayraktar, S., Gutierrez Barrera, A. M., Liu, D., Pusztai, L., Litton, J., Valero, V., Hunt, K., Hortobagyi, G. N., Wu, Y., Symmans, F., & Arun, B. (2013). USP-11 as a predictive and prognostic factor following neoadjuvant therapy in women with breast cancer. Cancer Journal, 19(1), 10–17. https://doi.org/10.1097/PPO.0b013e3182801b3a
Behrends, C., Sowa, M. E., Gygi, S. P., & Harper, J. W. (2010). Network organization of the human autophagy system. Nature, 466(7302), 68–76. https://doi.org/10.1038/nature09204
Chiang, S. Y., Wu, H. C., Lin, S. Y., Chen, H. Y., Wang, C. F., Yeh, N. H., Shih, J. H., Huang, Y. S., Kuo, H. C., Chou, S. J., & Chen, R. H. (2021). Usp11 controls cortical neurogenesis and neuronal migration through Sox11 stabilization. Sci Adv, 7(7). https://doi.org/10.1126/sciadv.abc6093
Dikic, I., & Elazar, Z. (2018). Mechanism and medical implications of mammalian autophagy. Nature Reviews Molecular Cell Biology, 19(6), 349–364. https://doi.org/10.1038/s41580-018-0003-4
Dong, S., Wang, Q., Kao, Y. R., Diaz, A., Tasset, I., Kaushik, S., Thiruthuvanathan, V., Zintiridou, A., Nieves, E., Dzieciatkowska, M., Reisz, J. A., Gavathiotis, E., D’Alessandro, A., Will, B., & Cuervo, A. M. (2021). Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature, 591(7848), 117–123. https://doi.org/10.1038/s41586-020-03129-z
Dwane, L., O’Connor, A. E., Das, S., Moran, B., Mulrane, L., Pinto-Fernandez, A., Ward, E., Blumel, A. M., Cavanagh, B. L., Mooney, B., Dirac, A. M., Jirstrom, K., Kessler, B. M., Ni Chonghaile, T., Bernards, R., Gallagher, W. M., & O’Connor, D. P. (2020). A functional genomic screen identifies the deubiquitinase USP11 as a novel transcriptional regulator of ERalpha in breast cancer. Cancer Research, 80(22), 5076–5088. https://doi.org/10.1158/0008-5472.CAN-20-0214
Galluzzi, L., & Green, D. R. (2019). Autophagy-independent functions of the autophagy machinery. Cell, 177(7), 1682–1699. https://doi.org/10.1016/j.cell.2019.05.026
Garcia, D. A., Baek, C., Estrada, M. V., Tysl, T., Bennett, E. J., Yang, J., & Chang, J. T. (2018). USP11 Enhances TGFbeta-induced epithelial-mesenchymal plasticity and human breast cancer metastasis. Molecular Cancer Research, 16(7), 1172–1184. https://doi.org/10.1158/1541-7786.MCR-17-0723
Georges, A., Marcon, E., Greenblatt, J., & Frappier, L. (2018). Identification and characterization of USP7 targets in cancer cells. Science and Reports, 8(1), 15833. https://doi.org/10.1038/s41598-018-34197-x
Han, T., Guo, M., Gan, M., Yu, B., Tian, X., & Wang, J. B. (2018). TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy, 14(12), 2035–2048. https://doi.org/10.1080/15548627.2018.1491493
Huang, Y. Y., Zhang, C. M., Dai, Y. B., Lin, J. G., Lin, N., Huang, Z. X., & Xu, T. W. (2021). USP11 facilitates colorectal cancer proliferation and metastasis by regulating IGF2BP3 stability. Am J Transl Res, 13(2), 480–496.
Jurga, M., Abugable, A. A., Goldman, A. S. H., & El-Khamisy, S. F. (2021). USP11 controls R-loops by regulating senataxin proteostasis. Nature Communications, 12(1), 5156. https://doi.org/10.1038/s41467-021-25459-w
Kaushik, S., & Cuervo, A. M. (2018). The coming of age of chaperone-mediated autophagy. Nature Reviews Molecular Cell Biology, 19(6), 365–381. https://doi.org/10.1038/s41580-018-0001-6
Li, X., Yang, K. B., Chen, W., Mai, J., Wu, X. Q., Sun, T., Wu, R. Y., Jiao, L., Li, D. D., Ji, J., Zhang, H. L., Yu, Y., Chen, Y. H., Feng, G. K., Deng, R., Li, J. D., & Zhu, X. F. (2021). CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy, 1-18. doi:https://doi.org/10.1080/15548627.2021.1912270
Liu, H., Liu, M., He, B., & Li, Q. (2021). Inhibition of USP11 sensitizes gastric cancer to chemotherapy via suppressing RhoA and Ras-mediated signaling pathways. Clin Res Hepatol Gastroenterol, 101779. doi:https://doi.org/10.1016/j.clinre.2021.101779
Liu, J., Xia, H., Kim, M., Xu, L., Li, Y., Zhang, L., Cai, Y., Norberg, H. V., Zhang, T., Furuya, T., Jin, M., Zhu, Z., Wang, H., Yu, J., Li, Y., Hao, Y., Choi, A., Ke, H., Ma, D., & Yuan, J. (2011). Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell, 147(1), 223–234. https://doi.org/10.1016/j.cell.2011.08.037
Pei, G., Buijze, H., Liu, H., Moura-Alves, P., Goosmann, C., Brinkmann, V., Kawabe, H., Dorhoi, A., & Kaufmann, S. H. E. (2017). The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy. Autophagy, 13(12), 2041–2055. https://doi.org/10.1080/15548627.2017.1376160
Platta, H. W., Abrahamsen, H., Thoresen, S. B., & Stenmark, H. (2012). Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. The Biochemical Journal, 441(1), 399–406. https://doi.org/10.1042/BJ20111424
Qiao, L., Zhang, Q., Sun, Z., Liu, Q., Wu, Z., Hu, W., Bao, S., Yang, Q., & Liu, L. (2021). The E2F1/USP11 positive feedback loop promotes hepatocellular carcinoma metastasis and inhibits autophagy by activating ERK/mTOR pathway. Cancer Letters, 514, 63–78. https://doi.org/10.1016/j.canlet.2021.05.015
Rabinowitz, J. D., & White, E. (2010). Autophagy and metabolism. Science, 330(6009), 1344–1348. https://doi.org/10.1126/science.1193497
Rong, Y., Fan, J., Ji, C., Wang, Z., Ge, X., Wang, J., Ye, W., Yin, G., Cai, W., & Liu, W. (2021). USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating Beclin 1. Cell Death and Differentiation. https://doi.org/10.1038/s41418-021-00907-8
Schuck, S. (2020). Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci, 133(17). https://doi.org/10.1242/jcs.246322
Sowa, M. E., Bennett, E. J., Gygi, S. P., & Harper, J. W. (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell, 138(2), 389–403. https://doi.org/10.1016/j.cell.2009.04.042
Sun, H., Wang, R., Liu, Y., Mei, H., Liu, X., & Peng, Z. (2021). USP11 induce resistance to 5-fluorouracil in colorectal cancer through activating autophagy by stabilizing VCP. Journal of Cancer, 12(8), 2317–2325. https://doi.org/10.7150/jca.52158
Ting, X., Xia, L., Yang, J., He, L., Si, W., Shang, Y., & Sun, L. (2019). USP11 acts as a histone deubiquitinase functioning in chromatin reorganization during DNA repair. Nucleic Acids Research, 47(18), 9721–9740. https://doi.org/10.1093/nar/gkz726
Wang, W., Wang, J., Yan, H., Zhang, K., & Liu, Y. (2019). Upregulation of USP11 promotes epithelialtomesenchymal transition by deubiquitinating Snail in ovarian cancer. Oncology Reports, 41(3), 1739–1748. https://doi.org/10.3892/or.2018.6924
White, E., & DiPaola, R. S. (2009). The double-edged sword of autophagy modulation in cancer. Clinical Cancer Research, 15(17), 5308–5316. https://doi.org/10.1158/1078-0432.CCR-07-5023
Yang, Q., Wang, R., & Zhu, L. (2019). Chaperone-mediated autophagy. Advances in Experimental Medicine and Biology, 1206, 435–452. https://doi.org/10.1007/978-981-15-0602-4_20
Yang, Y., & Klionsky, D. J. (2020). Autophagy and disease: unanswered questions. Cell Death and Differentiation, 27(3), 858–871. https://doi.org/10.1038/s41418-019-0480-9
Yang, Z., & Klionsky, D. J. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Current Opinion in Cell Biology, 22(2), 124–131. https://doi.org/10.1016/j.ceb.2009.11.014
Zalckvar, E., Berissi, H., Mizrachy, L., Idelchuk, Y., Koren, I., Eisenstein, M., Sabanay, H., Pinkas-Kramarski, R., & Kimchi, A. (2009). DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Reports, 10(3), 285–292. https://doi.org/10.1038/embor.2008.246
Zhang, L., Zhou, F., Drabsch, Y., Gao, R., Snaar-Jagalska, B. E., Mickanin, C., Huang, H., Sheppard, K. A., Porter, J. A., Lu, C. X., & ten Dijke, P. (2012). USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-beta type I receptor. Nature Cell Biology, 14(7), 717–726. https://doi.org/10.1038/ncb2522
Zhang, S., Xie, C., Li, H., Zhang, K., Li, J., Wang, X., & Yin, Z. (2018). Ubiquitin-specific protease 11 serves as a marker of poor prognosis and promotes metastasis in hepatocellular carcinoma. Laboratory Investigation, 98(7), 883–894. https://doi.org/10.1038/s41374-018-0050-7
Zhou, Z., Luo, A., Shrivastava, I., He, M., Huang, Y., Bahar, I., Liu, Z., & Wan, Y. (2017). Regulation of XIAP turnover reveals a role for USP11 in promotion of tumorigenesis. eBioMedicine, 15, 48–61. https://doi.org/10.1016/j.ebiom.2016.12.014
Funding
This work was supported by the National Natural Science Foundation of China (Grant no. 31530016, 32,090,031, 31,761,133,012, and 31,800,683) the National Basic Research Program of China (grant no. 2017YFA0503900), the Shenzhen Science and Technology Innovation Commission (grant no. JCYJ20180507182213033 and JCYJ20170412113009742).
Author information
Affiliations
Guangdong Key Laboratory for Genome Stability & Disease Prevention and Carson International Cancer Center, Marshall Laboratory of Biomedical Engineering, Shenzhen University School of Medicine, Shenzhen, 518060, Guangdong, China
Zheng Li, Shaohong Rao, Shuping Yuan, Bin Peng & Xingzhi Xu
College of Life Sciences, Capital Normal University, Beijing, 100048, China
Chunwei Song & Hongchang Zhao
Institute of Translational Medicine, College of Medicine, Qingdao University, Qingdao, 266000, Shandong, China
Min Zhu
Shenzhen University-Friedrich Schiller Universität Jena Joint PhD Program in Biomedical Sciences, Shenzhen University School of Medicine, Shenzhen, 518060, Guangdong, China
Shuping Yuan & Xingzhi Xu
Corresponding author
Correspondence to Xingzhi Xu.
Ethics declarations
Conflicts of interest
The authors declare no potential conflicts of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (PDF 340 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Z., Rao, S., Song, C. et al. Ubiquitin carboxyl-terminal hydrolase 11 promotes autophagy by de-ubiquitinating and stabilizing Beclin-1. GENOME INSTAB. DIS. 3, 47–55 (2022). https://doi.org/10.1007/s42764-022-00061-6
Received06 January 2022
Revised23 January 2022
Accepted25 January 2022
Published06 February 2022
Issue DateFebruary 2022
DOIhttps://doi.org/10.1007/s42764-022-00061-6
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable linkKeywords
USP11
Beclin-1
Autophagy
De-ubiquitination
Class III phosphatidylinositol 3-kinase complex








用户登录
还没有账号?
立即注册