MicroRNA-mediated reprogramming of glucose, fatty acid and amino acid metabolism in cancer
Review Article
Genome Instability & Disease (2022)
Cancer cells possess a unique metabolic phenotype, rewiring their metabolic pathways to support energy-intensive and biosynthetic requirements for exuberant proliferation and migration. Attention has focused on the role of microRNAs (miRNAs) in mediating metabolic shifts in cancer cells. miRNAs participate in cancer metabolism reprogramming mainly by directly silencing the expression of specific genes. This review focuses on miRNA regulation of cancer metabolic reprogramming, including glucose, fatty acid and amino acid metabolism, and allows a deeper understanding of importance of miRNAs in tumorigenesis and progression, therefore, providing new avenues for cancer therapy strategy. Our understanding of the molecular mechanisms underlying cancer cell metabolism and their functions at various stages of tumorigenesis has increased due to the development of new biochemical and biological tools (Benjamin et al., 2012). The remodelling of cellular metabolism represents a fascinating hallmark of cancer. Cancer cells rewire, mutationally activate and transcriptionally upregulate multiple metabolic pathways, leading to aberrant production or consumption of essential biomolecules such as glucose, amino acids, nucleotides and lipids that in turn fuel tumour growth and malignancy (Pavlova & Thompson, 2016). Cancer cells preferentially execute the “Warburg effect”, that is, enhanced anaerobic glycolysis, even when there is sufficient oxygen supply (Hanahan, 2022). The metabolic traits associated with tumour cells are also crucial for allowing them to adjust to changes in the oxygen and nutrients available in the tumour microenvironment, sustaining proliferation and resisting apoptosis (Wellen & Thompson, 2012). In addition, deregulated anabolism/catabolism of metabolites, particularly glutamine, serine and glycine, act as metabolic regulators, supporting cancer cell growth (Li & Zhang, 2016). Oncogenes, tumour-suppressor genes and miRNA genes participate in a multistep sequential alteration process in multiple cancer cells (Croce, 2008). Moreover, numerous miRNA-related studies have confirmed the roles of small non-coding RNAs in fine-tuning gene expression at different stages of tumorigenesis (Goodall & Wickramasinghe, 2021). miRNAs are short non-coding RNAs (ncRNAs) approximately 18–25 nucleotides that are present in all eukaryotic cells (Ha & Kim, 2014) and regulate posttranscriptional gene expression by binding to the 3’ untranslated region (3’UTR) of messenger RNAs (mRNAs) (Gebert & MacRae, 2019). Since the first miRNA was identified in 1993 (transcribed from the Caenorhabditis elegans lin-4 locus) (Lee et al., 1993), studies have shown that miRNAs are involved in multiple important biological processes, including cell proliferation and differentiation (Shenoy & Blelloch, 2014), apoptosis (Gorbea et al., 2017), senescence (Jauhari et al., 2022), autophagy (Gibbings et al., 2012), metastasis (Zhang et al., 2015a), and immune responses (Seeley et al., 2018). Moreover, miRNAs maintain chromatin structure and metabolic homeostasis (Gaal, 2021). Due to their extensive roles, miRNAs are a novel class of essential gene regulators, which are related to various pathological conditions, including metabolic diseases and associated cancers (Bracken et al., 2016; Rottiers & Naar, 2012). Here we focus on the regulation of glucose, fatty acid and amino acid metabolism in human cancers and review a detailed look at studies on the roles of different miRNAs in cancer metabolism by targeting key enzymes, transporter proteins or nuclear receptors (NRs). To further understand the importance of miRNAs in tumorigenesis and progression, thus providing new approaches for cancer therapy strategies. A typical feature shared by almost all cancer cells is enhanced glucose uptake and aerobic glycolysis (Hanahan & Weinberg, 2011). The first rate-limiting step in glucose metabolism is glucose transport across plasma membranes of cancer cells, mediated by facilitative glucose transporters (GLUTs) (Macheda et al., 2005). Deregulation of GLUTs enhances glucose uptake, accelerates metabolism and increases glucose requirement for tumorous tissues. The altered expression and activity of key GLUTs in cancer cells is driven by multiple factors, particularly miRNAs (Table 1). The GLUTs isoforms GLUT1-GLUT4 were investigated most intensively with a particular focus on GLUT1. In breast cancer, miR-140-5p is downregulated and suppresses glycolysis by directly targeting GLUT1, resulting in an anti-glycolytic and anti-proliferative effect, while the ectopic expression of miR-22 inhibits cell proliferation and invasion by targeting GLUT1 (Chen et al., 2015; He et al., 2019). miR-340 inhibits cell proliferation and induces apoptosis by targeting GLUT1 partly through regulating PCNA, Bax expression and PI3K/AKT pathway in bladder cancer (Xu et al., 2021a). GLUT1 is also a glycolytic target that is modulated by miR-328 in colon cancer and osteosarcoma (Santasusagna et al., 2018; Yi et al., 2020), by miR-132 and miR-378a in prostate cancer (Cannistraci, 2022; Qu et al., 2016), by miR-1291 in renal cell carcinoma (RCC) (Yamasaki et al., 2013), by miR-148a in intrahepatic cholangiocarcinoma (Tiemin et al., 2020), by miR-148b in gastric cancer (GC) (Ding et al., 2017), by miR-199a-5p in non-small cell lung cancer (NSCLC) (Xu et al., 2022), by miR-144 in ovarian cancer (Fan et al., 2016a) and by miR-192-5p in hepatocellular carcinoma (HCC) (Gu et al., 2020). In addition to GLUT1, the expression of GLUT3 was suppressed by miR-195-5p in bladder cancer (Fei et al., 2012) and by miR-106a in glioblastoma (GBM) (Dai et al., 2013), resulting in the inhibition of cell proliferation and glucose uptake. In HDAC2 knockdown GBM, miR-3189 mediated GLUT3 inhibition shows an anti-tumorigenic effect and cell death by regulating glucose metabolism (Kwak et al., 2022). Unlike GLUT1 or GLUT3, GLUT4 is a glucose carrier entirely dependent upon insulin. miR-150 is reported as a negative regulator of GLUT4 in insulin-resistant cardiomyocytes (Ju et al., 2020), resulting in the inhibition of glucose uptake. Whether the expression level of GLUT4 is related to the metabolic phenotype of cancer remains to be investigated. Table 1 Summary of metabolic enzymes and transporters involved in cancer metabolic reprogramming directly regulated by miRNAs Glycolysis (Fig. 1a) occurs after glucose is transported into cancer cells, facilitating the conversion of glucose molecules into macromolecular precursors required for the pentose phosphate pathway as well as for lipid, amino acid and nucleotide biosynthesis (Vander Heiden et al., 2009). The glycolytic pathway involves catabolism of glucose to pyruvate in 9–10 biochemical steps (Li & Zhang, 2016). Hexokinases (HKs) phosphorylates glucose to glucose-6-phosphate; Phosphoglucose isomerase (PGI) catalyses the interconversion between glucose-6-phosphate and fructose-6-phosphate; Phosphofructokinase (PFK) is the main rate-limiting enzyme that converts fructose-6-phosphate to fructose-1,6-bisphosphate; Aldolase A (ALDOA) converts fructose-1,6-bisphosphate into glyceraldehyde-3-phosphate and dihydroxyacetone phosphate; Triosephosphate isomerase 1 (TPI1) catalyses the isomerisation of glyceraldehyde-3-phosphate and dihydroxyacetone phosphate; Phosphoglycerate kinase 1 (PGK1) catalyses the conversion of 1,3-diphosphoglycerate to 3-phosphoglycerate, phosphoglycerate mutase 1 (PGAM1) catalyses the interconversion of 3-phosphoglycerate and 2-phosphoglycerate; Enolase 1 (ENO1) converts 2-phosphoglycerate to phosphoenolpyruvate; and pyruvate kinase M2 (PKM2) is the last rate-limiting enzyme that catalyses the final glycolytic step, converting phosphoenolpyruvate to pyruvate. Additionally, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is associated with PKM2, catalyses the reversible oxidative phosphorylation of glyceraldehyde-3-phosphate in the presence of inorganic phosphate and nicotinamide adenine dinucleotide (NAD). Lactate dehydrogenase (LDH), a critical enzyme, transforms pyruvate into lactate in the final step of anaerobic glycolysis. Lactate is transported out of the cells by monocarboxylate transporters (MCTs) where it is converted into glucose and recycled back to the tumour cells. MiRNAs-mediated regulation of Glucose and Fatty Acid Metabolism in Cancer. Metabolic pathways in this schematic diagram include a-d glucose, and e fatty acid metabolism. Glucose metabolism contains a glycolysis and c pentose phosphate pathway (PPP) in the cytoplasm and b Krebs tricarboxylic acid (TCA) cycle in the mitochondrion. Glucose transported into the cells by GLUTs, undergoes glycolysis to generate pyruvate which then goes through TCA cycle to generate ATP. In cancer cells, the flux of pyruvate entering TCA cycle decreases and the majority of pyruvate undergoes lactic acid fermentation for the rapid production of ATP. The PPP branches after the first step of glycolysis. In cancer cells, increased PPP supplies the ribose sugar for the nucleotide synthesis necessary for rapidly growing cells. The enzymes are denoted by ovals. Enzymes in the glycolysis, PPP, TCA cycle and d glycogenesis are respective shaded in blue, green, yellow and brown. cancer cells e fatty acid metabolism contains de novo fatty acid synthesis and exogenous uptake. In cancer cells, fatty acid synthesis uses the substrate acetyl-CoA, which can be generated from glucose, glutamine, or acetate. The product of de novo fatty acid synthesis is palmitate, which serves as a precursor to other fatty acids (via SCD). Exogenous FA can be brought into cells via specialized transporter, including CD36. Cholesterol carried by LDL particles can be taken up via the LDLR. Excess cholesterol is exported to the blood by ABCA1 or the homodimer of ABCG1. Enzymes for individual chemical reactions are labelled as ovals and denoted in purple next to the arrows connecting two metabolites. G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F-1,6-BP, fructose-1,6-bisphosphate; GA3P, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; 1,3BPG, 1,3-disphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; α-KG, α-ketoglutarate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; Xu5P, xylulose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrose-4-phosphate; PRPP, phosphoribosyl pyrophosphate; G1P, glucose-1-phosphate; UDPG, UDP-glucose; FA, fatty acid; FA-CoA, fatty acid-CoA; LDL, low-density lipoprotein; Pathway enzymes are: GLUTs, glucose transporters; HKs, hexokinases; GPI, phosphoglucose isomerase; PFKs, phosphofructokinases; ALDOA, aldolase A; TPI1, triosephosphate isomerase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGK1, phosphoglycerate kinase 1; PGAM1, phosphoglycerate mutase 1; ENO1, enolase 1, PKM2, pyruvate kinase M2; LDHA, lactate dehydrogenase A; MCTs, monocarboxylate transporters; MPC, mitochondrial pyruvate carrier; PDH, pyruvate dehydrogenase; PDKs, pyruvate dehydrogenase kinases; CS, citrate synthase; ACO2, aconitase 2; IDH2, isocitrate dehydrogenase 2; OGDH, oxoglutarate dehydrogenase; SCS, succinyl-CoA synthetase; SDH, succinate dehydrogenase; FH, fumarate hydratase; MDH1, malate dehydrogenase 1; G6PD, glucose-6-phosphate dehydrogenase; PGLS, 6-phosphogluconolactonase; 6PGD, 6-phosphogluconate dehydrogenase; RPIA, ribose-5-phosphate isomerase A; RPE, ribulose-5-phosphate epimerase; PRPS1, phosphoribosyl pyrophosphate synthetase 1; TKT, transketolase; TALDO, transaldolase; PGM1, phosphoglucomutase 1; UGP2, UDP-glucose pyrophosphorylase 2; GYG1, glycogenin 1; GYS, glycogen synthase; GBE, glucan branching enzyme; GSK-3β, glycogen synthase kinase-3 beta; CIC, citrate carrier; ACLY, ATP-citrate lyase; ACSS2, acyl-CoA synthetase short chain family member 2; ACC1/2, acetyl-CoA carboxylase alpha/beta; FASN, fatty acid synthase; SCD, stearoyl-CoA desaturase; CPT, carnitine palmitoyltransferase; CACT, carnitine acylcarnitine translocase; LDLR, low-density lipoprotein receptor; ABCA1, ATP-binding cassette subfamily A member 1; ABCG1, ATP-binding cassette subfamily G member 1 miRNAs directly regulate key enzymes involved in glycolysis in response to tumour needs. HKs, which exist four major isoforms: HK1-HK4, are the first rate-limiting enzyme in aerobic glycolysis. HK2, which is predominant overexpression in cancer, is essential for tumour metabolism. Increased expression of HK2 plays an important role in tumour growth, metastasis and tumorigenesis. Multiple regulatory axes of HK2 expression have been identified in various cancer cell types, including miR-34a in colorectal cancer (CRC) cells (Kim et al., 2013), miR-125a/b and miR-143 in NSCLC cells (Fang et al., 2012), miR-202 and miR-199a-5p in liver cancer cells (Guo et al., 2015; Wang et al., 2020a; Zhang et al., 2015b), miR-125b in bladder cancer cells (Liu et al., 2020a), miR-148 in cervical cancer cells (Yang et al., 2021) and miR-603 in ovarian cancer cells (Lu et al., 2019). It is remarkable that miR-143 direct targets HK2 and inhibits glucose metabolism in head and neck squamous cell carcinoma (HNSCC)-derived cell lines (Peschiaroli et al., 2013), breast cancer cells (Jiang et al., 2012), NSCLC cells (Fang et al., 2012), prostate cancer cells (Zhou et al., 2015), RCC cells (Yoshino et al., 2013), oral squamous cell carcinoma (OSCC) cells (Sun & Zhang, 2017), gallbladder cancer (GBC) cells (Chen et al., 2019a) and colon cancer cells (Gregersen et al., 2012). Taken together, miR-143-HK2 axis plays an important role in tumorigenesis and represents a potential cancer therapeutic strategy. Except for HK2, other enzymes in glycolysis are regulated by miRNAs as well. GPI plays an important role in glycolysis and gluconeogenesis and is associated with cancer progression and poor prognosis. miR-34a has been also shown to repress GPI expression in CRC cells (Kim et al., 2013). PFK is the second rate-limiting enzyme involved in glycolysis. Three PFK isozymes exist in humans: muscle (PFKM), liver (PFKL) and platelet (PFKP). The miR-338-PFKL axis regulates the Warburg effect and anti-tumorigenesis in HCC cells treated with 125I irradiation and provides a new potential strategy for HCC clinical treatment (Zheng et al., 2019). PFKL is also directly targeted by miR-185-5p and miR-128 in lung cancer cells (Li et al., 2021a; Yang et al., 2016a). miR-520a/b/e are major regulators of glycolysis by directly targeting the 3’ UTR of PFKP in HCC cells (Park et al., 2013). ALDOA is a glycolytic target that is modulated by miR-122 in colon cancer, pancreatic cancer and bile duct carcinoma cells (Cui et al., 2019; Li et al., 2019a; Xu et al., 2018a). ALDOC, another member of the aldolase family mainly in brain, is targeted by let-7 in breast cancer cells (Reinsborough et al., 2021). TPI downregulation is regulated by miR-22 and miR-28 with implications in tumorigenesis in CRC cells (Lone et al., 2018). PGK1 is an important enzyme in the pathway of metabolic glycolysis, which has been reported to regulate malignancies, such as CRC, breast cancer, and liver cancer. miR‐548c‐5p is a useful tumour suppressor in CRC by targeting PGK1 (Ge et al., 2019). PGK1 is also targeted by miR-143 in prostate cancer (Cao et al., 2017), by miR-450b-3p in HCC (Chen et al., 2019b), by miR-16-1-3p in breast cancer (Ye et al., 2020a), and by miR-6869-5p in glioma (Wang et al., 2020b). PGAM1 is a functional target for miR-3614-5p. miR-3614-5p-PGAM1 axis regulates the malignant phenotype through activating the TGF-β signalling pathway in NSCLC cells (Li et al., 2020). miR-22-3p targets ENO1, regulating the proliferation of retinoblastoma cells (Liu et al., 2018). PKM2, which is the isoform of pyruvate kinase expressed by tumours, has emerged as a critical regulator of the glycolytic process during cancer development (Sun et al., 2011) and is a well-known miRNA target in cancer cells. Several miRNA-PKM2 axes have been shown to play a role in glycolysis, proliferation, apoptosis, metastasis and chemotherapy resistance in different cancers, for example, the miR-625-5p-PKM2 axis in melanoma cells (Zhang et al., 2019a), the miR-139-5p-PKM2 axis in gallbladder carcinoma cells (Chen et al., 2018a), the miR-326-PKM2 axis in glioma and thyroid cancer cells (Kefas et al., 2010; Yu et al., 2018), the miR-199a-5p-PKM2 and miR-491-5p-PKM2 axes in HCC cells (Xu et al., 2018b; Zhang et al., 2015b), the miR-489-3p-PKM2 axis in pancreatic cancer cells (Zhang et al., 2021a), the c-Myc-miR-184-PKM2 axis in clear-cell renal cell carcinoma (ccRCC) cells (Huang et al., 2016) and the regulatory circuit of PKM2-NF-κB-miR-148a/152 in triple-negative breast cancer (TNBC) cells (Xu et al., 2015). GAPDH is used extensively to normalize gene expression data. Interestingly, GAPDH is a key enzyme in aerobic glycolysis that plays an essential role in tumour metabolism. A recent report showed that miR-644a expression suppresses the Warburg effect by directly targeting GAPDH expression (Ebron et al., 2019; Sikand et al., 2012), and some predicted miRNA-GAPDH regulatory axes in cervical cancer need to be verified (Liu et al., 2020b). Most pyruvate molecules are reduced to lactate during glycolysis by enzyme LDH in cancer cells. LDHA has an aberrantly high expression in multiple cancers and might be involved in tumour progression and metabolism by regulated Warburg effect. LDHA is directly targeted by different miRNAs in different cancer cells, including miR-30d-5p in GBC cells (He et al., 2018a), miR-30a-5p and miR-34a in breast cancer cells (Li et al., 2017a; Xiao et al., 2016), miR-383 in ovarian cancer cell (Han et al., 2017), miR-449a in NSCLC cells (Li et al., 2018), miR-142-3p in HCC cells (Hua et al., 2018), miR-204-3p in bladder cancer cells (Guo et al., 2019), miR-329-3p and miR-323a-3p in osteosarcoma cells (Chen et al., 2018b; Li et al., 2021b) and miR-489-3p in pancreatic cancer cells (Zhang et al., 2021a). Lactate is then secreted outside of the cells by MCTs. MCTs, especially MCT1 and MCT4, are high expression in human cancers. Studies showed that MCTs are also under the control of miRNAs. For example, MCT1 expression can be suppressed by targeting miR-124 in medulloblastoma and breast cancer cells (Hou et al., 2019; Li et al., 2009), targeting miR-342-3p in HCC and TNBC cells (Komoll et al., 2021; Romero-Cordoba et al., 2018) and targeting miR-449a in glioma cells (Sheng et al., 2020), while MCT4 is targeted by miR-145-5p in HCC cells (Zhao et al., 2019a). The rapid glycolytic process in cancer cells leads to pyruvate shunting from the mitochondria to the cytosol where it is converted to lactate (Vaupel & Multhoff, 2021). Under aerobic conditions, the pyruvate produced by the glycolysis pathway is transported to the mitochondria and used in the Krebs tricarboxylic acid (TCA) cycle (Fig. 1b). Because oxygen is required for the regeneration of NAD+ and flavin adenine dinucleotide (FAD), the TCA cycle proceeds in the mitochondria only under aerobic conditions (Akram, 2014). In humans, mitochondrial pyruvate carrier (MPC) delivers pyruvate from the mitochondrial intermembrane space to the mitochondrial matrix (Bricker et al., 2012). No major cancer-associated miRNAs are known to regulate MPC genes. After passing through MPC, pyruvate is converted to acetyl-CoA by the pyruvate dehydrogenase complex (PDC), serving as a crossroad between cytoplasmic glycolysis and the mitochondrial TCA cycle. PDC, a nuclear-encoded mitochondrial multienzyme complex, comprises the following three catalytic enzymes and their regulator proteins: pyruvate dehydrogenase (PDH), dihydrolipoamide S-acetyltransferase (DLAT) and dihydrolipoamide dehydrogenase (DLD). PDH activity is inhibited through phosphorylation by pyruvate dehydrogenase kinase (PDK) (Wang et al., 2021). Cancer cells rely more on aerobic glycolysis and less on the TCA cycle, ensuring that cells generate multiple biosynthesis materials while providing abundant ATP (Gray et al., 2014), yet there is increasing evidence showing that miRNAs regulate mitochondrial metabolic processes during cancer progression (Bienertova-Vasku et al., 2013; Tomasetti et al., 2014). The TCA cycle itself operates in the mitochondrial matrix and is an amphibolic pathway that plays an important role in the integration of multiple catabolic and anabolic pathways such as glycolysis and gluconeogenesis. There are a total of eight steps in the TCA cycle, three of which are irreversible as follows: the generation of citrate from oxaloacetate and acetyl-CoA, the conversion of isocitrate to alpha-ketoglutarate (α-KG) and the formation of succinyl-CoA from α-KG (Anderson et al., 2018). Citrate synthase (CS) catalyses the synthesis of citrate from oxaloacetate and acetyl-CoA, aconitase (ACO), catalyses the conversion of citrate to isocitrate, isocitrate dehydrogenase (IDH) catalyses the oxidative decarboxylation of isocitrate to α-KG, α-KG dehydrogenase complex (KGDHC) catalyses the overall conversion of α-KG to succinyl-CoA, succinyl-CoA synthetase (SCS) catalyses the reversible conversion between succinyl-CoA and succinate, succinate dehydrogenase (SDH) converts succinate to fumarate, fumarate hydratase (FH) converts fumarate to malate and malate dehydrogenase (MDH) catalyses the NAD/NADH-dependent reversible oxidation of malate to oxaloacetate. PDKs have PDK1-PDK4 isoforms in mammals, which are responsible for tissue- or cell-specific regulation of PDH. PDK1 is involved in the physiological regulation of transcription regulation, protein synthesis, cell proliferation, migration and apoptosis, and may be a viable cancer biomarker. PDK1 is targeted by let-7g, leading to the regulation of aerobic glycolysis and cancer progression via mTOR signalling pathway in HCC cells (Ma et al., 2014). It is also targeted by miR-34a in CRC cells (Kim et al., 2013), by miR-155-5p in cervical cancer cells (Wang et al., 2018), by miR-9 in prostate cancer cells (Sheng et al., 2021), and miR-128-3p-targeted PDK1 contributes to mitochondrial dysfunction and apoptosis in glioma cells (Qu et al., 2020a). In GC cells, miR-422a suppresses PDK2 to restore activity of PDH, providing a promising therapeutic target for anti-tumour treatment. Importantly, miR-422a-PDK2 axis also has been shown to influence de novo lipogenesis, and it subsequently affects reactive oxygen species (ROS) and retinoblastoma protein (RB) phosphorylation levels, resulting in G1 phase arrest (He et al., 2018b). PDK2 is also targeted by miR-149-3p in CRC cells (Liang et al., 2020 ), and by miR-214 in HCC cells (Yu, 2019). miR-497-5p inhibits GC cell proliferation and growth via directly targeting and suppressing the expression of PDK3 (Feng et al., 2019). PDK4 expression can be regulated by miR-23a in CRC cells (Deng et al., 2018), miR-182 in lung cancer cells (Li et al., 2017b), miR-9-5p in HCC cells (Si et al., 2021), miR-5683 in GC cells (Miao et al., 2020), and miR-16-5p in cervical cancer cells (Zhao et al., 2020). In liver, skeletal muscle and heart tissues, the expression of PDK4 is also regulated by ERRs (estrogen-related receptors) and HNF4α (hepatocyte nuclear factor 4α) (Ma et al., 2005; Wende et al., 2005), two subfamilies of NRs. NRs are a superfamily of ligand-activated transcription factors that control the expression of a large number of genes involved in the regulation of a variety of pathways, including glucose and fatty acid metabolism (Weikum et al., 2018). ERRs, a subfamily of orphan NRs that comprise ERRα, ERRβ, and ERRγ, are well known as central transcriptional regulators of energy homeostasis and mitochondrial biogenesis (Ranhotra, 2018). ERRα is a miR-125a target gene in OSCC cells (Tiwari et al., 2014), a miR-137 target gene in nasopharyngeal carcinoma cells (Liu et al., 2022), and a miR-497 target gene in ERα negative breast cancer cells (Han et al., 2016). ERRγ is targeted by miR-320a in breast cancer cells (Lu et al., 2015) and by miR-940 in HCC cells (Yuan et al., 2015). PGC-1α, first found as a coactivator of PPARγ (peroxisome proliferator-activated receptor γ), is the most notable and potent coactivator of ERRα, is also regulated by miRNAs. miR-23a downregulates PGC-1α in HCC cells (Wang et al., 2012) while miR-217 interacts with PGC-1α mRNA in breast cancer cells (Zhang et al., 2017). HNF4α is another subfamily of NRs that is involved in the regulation of a variety of pathways, including glucose, lipid, steroid, and amino acid metabolism in the liver (Schrem et al., 2002). In HCC cells, HNF4α is directly regulated by a few of miRNAs, including miR-21, miR-24, miR-629, miR-34a, miR-34c-5p and miR-449a (Hatziapostolou et al., 2011; Ning et al., 2014; Wang & Burke, 2013). Next, miRNAs that target CS during cancer development have not been described. Combined AGO-PAR-CLIP and RNA-seq analyses identified ACO2 as a prominent miR-450a target in ovarian cancer (Muys et al., 2019). ACO2 is targeted by miR-744-5p in spindle cell oncocytoma, one rare tumour accouting for 0.1–0.4% of all sellar tumours (Krokker et al., 2019). miR-183 has been shown to directly target IDH2 in GBM cells (Tanaka et al., 2013). KGDHC is a rate-limiting enzyme of the TCA cycle and has three components including α-KG dehydrogenase (OGDH) or OGDHL, dihydrolipoamide S-succinyltransferase (DLST), and DLD. OGDHL, a candidate tumour suppressor that plays a pivotal role in mitochondrial metabolism, participates in the miR-214/TWIST1 negative feedback loop in pancreatic cancer cells (Liu et al., 2019a). SUCLG2 is regulated by miR-155-5p in RCC-derived cell lines (Boguslawska et al., 2019). SDH complex is a key respiratory enzyme composed of six subunits encoded by SDHA, SDHB, SDHC, SDHD, SDHAF1 and SDHAF2, the last two of them coding for associated accessory factors. SDHB is a miR-142-5p target in CRC cells and facilitates aerobic glycolysis (Liu et al., 2017), and a miR-96-3p target in papillary thyroid cancer cells and influences the downstream molecule AKT and mTOR (Zhao et al., 2019b). miR-210 targets SDHD with important consequences on the modulation of HIF1 activity and cell metabolism in NSCLC (Puissegur et al., 2011). No major cancer-associated miRNAs are known to directly regulate FH activity. Two MDH isozymes exist in humans: one is found in the cytoplasm (MDH1), and the other in the mitochondrial matrix (MDH2). They play pivotal roles in the malate-aspartate shuttle that operates in the metabolic coordination between cytosol and mitochondria and also under the controlled of miRNAs. Recent evidence has shown that miR-126-5p suppresses the enzymatic activity of MDH1 in NSCLC cells (Lima Queiroz et al., 2018). The pentose phosphate pathway (PPP) (Fig. 1c) was among the first metabolic pathways to be discovered and is considered important for tumorigenesis as it not only provides NADPH that fuels fatty acid synthesis and sustains the antioxidant response, but also supplies ribose for nucleotide synthesis (Patra & Hay, 2014). Glucose-6-phosphate dehydrogenase (G6PD) is the rate-limiting enzyme in the oxidative PPP, controlling the flux of glucose-6-phosphate into the pathway. G6PD catalyses the conversion of glucose-6-phosphate to 6-phosphogluconolactone, which is accompanied by NADPH production. 6-phosphogluconolactonase (PGLS) hydrolyses 6-phosphogluconolactone to produce 6-phosphogluconate. 6-phosphogluconate dehydrogenase (6PGD) converts 6-phosphogluconate to ribulose-5-phosphate, generating NAPDH. Phosphoribosyl pyrophosphate synthetase 1 (PRPS1) catalyses the phosphoribosylation of ribose-5-phosphate, forming 5-phosphoribosyl-1-pyrophosphate, which is necessary for purine metabolism and nucleotide biosynthesis. In non-oxidative PPP, ribose-5-phosphate isomerase A (RPIA) converts ribulose-5-phosphate to ribose-5-phosphate, and ribulose-5-phosphate epimerase (RPE) catalyses the reversible reaction that converts ribulose-5-phosphate to xylulose-5-phosphate. Transketolase (TKT) reversibly converts ribose-5-phosphate to sedoheptulose-7-phosphate and plays a role in channelling excess sugar phosphates to glycolysis in the PPP. Therefore, TKT can bi-directionally regulate carbon flux between non-oxidative PPP and glycolysis or gluconeogenesis. Transaldolase 1 (TALDO1) catalyses the conversion of sedoheptulose-7-phosphate to fructose-6-phosphate. miRNAs also regulate key enzymes in the PPP. G6PD, the first rate-limiting enzyme in the oxidative branch of PPP, is often activated in human malignancies to generate precursors for nucleotide and fatty acid synthesis. Studies have demonstrated that G6PD, 6PGD and TKT are endogenous targets of miR-1 and miR-206 in both cancer and non-neoplastic fibroblast cells (Singh et al., 2013). The miR-206-G6PD axis also play a role in HCC, rhabdomyosarcoma, nasopharyngeal carcinoma, and HR-HPV-positive cervical cancer cells (Coda et al., 2015; Cui et al., 2018; Guo et al., 2020; Wang et al., 2021a). G6PD is still regulated by miR-122 in HCC cells, by miR-613 in esophageal squamous cell carcinoma (ESCC) cells (Barajas et al., 2018; Su et al., 2020). miR-206 and miR-613 have rewired cellular metabolism by targeting 6PGD in ovarian and lung cancer cells (Zheng et al., 2017). miR-124 inhibits the proliferation and DNA synthesis of CRC cells by targeting PRPS1 and RPIA (Qiu et al., 2015). miR-497-TKT axis modulates the glutathione (GSH) and ROS levels, which subsequently promote cisplatin chemoresistance in cervical cancer (Yang et al., 2016b). However, no major cancer-associated miRNAs that regulate the enzymes PGLS, RPE, and TALDO1 have been identified. Excess glucose that is not consumed during normal metabolism is converted to glycogen through the glycogen synthesis pathway (Fig. 1d) and stored for later use (Liu et al., 2021). Glycogen synthesis pathway starts with the conversion of glucose-6-phosphate to glucose-1-phosphate by phosphoglucomutase-1 (PGM1). UDP-glucose pryophosphorylase 2 (UGP2) converts glucose-1-phosphate to UDP-glucose. Glycogenin 1 (GYG1) is a glycosyltransferase that catalyses the formation of a short glucose polymer from uridine diphosphate glucose. Glucan branching enzyme (GBE1) catalyses the branching of glycogen chains. Glycogen synthase (GYS) catalyses the addition of glucose monomers to growing glycogen molecules. GYG1 has been identified as a target of miR-194 in HCC cells (Morimoto et al., 2017). miR-564 inhibits the proliferation and invasion of breast cancer cells by directly targeting GYS1 (Mutlu et al., 2016). GYS1 is also a target gene of miR-140-5p in HepG2 cells (Li et al., 2021c). However, no major cancer-associated miRNAs are known to regulate PGM1, UGP2, and GBE1. Glycogen synthase kinase-3 beta (GSK-3β) is also involved in glycogen synthesis (Cohen, 1999). miRNAs target GSK-3β during some pathological processes, including miR-1275-mediated signalling in lung adenocarcinoma (Jiang et al., 2020), miR-27a-3p-mediated signalling in TNBC (Wu et al., 2020a), miR-26a-mediated signalling in cholangiocarcinoma (Zhang et al., 2012a), miR-224-mediated signalling in CRC (Li et al., 2016), miR-501-5p-mediated signalling in GC (Fan et al., 2016b), miR-769-mediated signalling in melanoma (Qiu et al., 2016), and miR-1246-mediated signalling in liver cancer stem cells (Chai et al., 2016). Cancer cells require large quantities of lipids and cholesterol. These are obtained through either taking up additional exogenous lipids and lipoproteins or activating de novo lipogenesis and cholesterol biosynthesis (Fig. 1e) (Currie et al., 2013). A sufficient supply of lipids is vital for survival and proliferation of cancer cells growing under unfavourable environments (Koundouros & Poulogiannis, 2020). Some key regulators of lipogenesis such as sterol regulatory element binding proteins (SREBPs), acetyl-CoA carboxylases (ACCs), fatty acid synthase (FASN), and stearoyl-CoA desaturase (SCD) are significantly upregulated in various human cancers (Rohrig & Schulze, 2016). A critical step in glucose-mediated de novo lipid synthesis is the release of citrate from mitochondria into the cytoplasm. Solute carrier family 25 member 1 (SLC25A1), also known as citrate carrier (CIC), functions as a key transporter in this process, providing a key precursor for both fatty acid and cholesterol synthesis (Convertini et al., 2016). However, the miRNA-mediated regulation of SLC25A1 has not been found. Lipid production depends on the cytosolic acetyl-CoA pool that is supplied by the following two metabolic enzymes: ATP citrate lyase (ACLY) and acyl-CoA synthetase short chain family member 2 (ACSS2) (Butler et al., 2020), particularly under nutrient-poor conditions. ACLY cleaves citrate into oxaloacetate and acetyl-CoA, while ACSS2 ligates acetate and coenzyme A to produce acetyl-CoA. miR-146b-5p directly targets the 3' UTR of ACLY in salivary adenoid cystic carcinoma (SACC) cells (Xie, 2020), while miR-22 regulates ACLY in breast cancer cells (Table 1) (Koufaris et al., 2016). miR-15a-5p was shown to inhibit ACSS2 expression in lung cancer cells (Ni et al., 2020). There are two isoforms of ACC in human genome, ACC1 and ACC2. ACC1 is located in the cytosol and is highly expressed in lipogenic tissues, while ACC2 is imbedded in the mitochondrial membrane and primarily located in oxidative tissues. ACC1 catalyse ATP-dependent carboxylation of acetyl-CoA following the conversion of citrate and acetate to acetyl-CoA, generating malonyl-CoA, which is a rate-limiting step in fatty acid synthesis (Zu et al., 2013). Malonyl-CoA made by ACC2 is thought to inhibit CPT1 (carnitine palmitoyltransferases 1), thus preventing fatty acid degradation. A recent study found that miR-206 inhibits HCV proliferation through depressing ACC1 lipid synthesis signalling pathway (Zheng, 2022). ACC1 is also targeted by miR-195 in breast cancer cells (Singh et al., 2015). FASN, a key lipogenic enzyme catalysing the last step in the de novo biogenesis of fatty acids, has been studied extensively in various cancers (Jones & Infante, 2015). miR-195 directly targets FASN in meningioma and breast cancer cells (Singh et al., 2015; Song et al., 2020), miR-4310 inhibits lipid synthesis by targeting FASN in HCC cells (Li et al., 2021d), and miR-497-5p inhibits progression of cervical cancer cells through targeting FASN (Zhang, 2022). SCD is an ER-resident integral membrane protein that catalyses the formation of the monounsaturated fatty acids oleic acid (18:1) and palmitoleic acid (16:1) from stearoyl-(18:0) and palmitoyl-CoA (16:0), respectively (Bai et al., 2015). SCD is regulated by several miRNAs in cancer cells, including miR-19b-1, miR-142, miR-544a, and miR-215 in CRC cells (Cruz-Gil et al., 2018; Xu et al., 2020a), miR-433-3p in nasopharyngeal carcinoma cells (Yin et al., 2021), and miR-4310 in HCC cells (Li et al., 2021d). In addition to de novo synthesis, lipid uptake from the exogenous environment is another important route through which cancer cells acquire fatty acids, with CD36 playing a critical role in this process (Pascual et al., 2017). CD36 is targeted by different species of miRNA that modulate its expression at a post-transcriptional level in a tissue-specific manner. An miR-1254-CD36 cascade exists in OSCC cells (Chen et al., 2021a; Chen et al., 2019c), while an miR-145-5p-CD36 cascade exists in breast cancer cells (Yuan et al., 2021). Another important transport process in lipogenesis is the importation of acyl-CoA into the mitochondria by the following three acyltransferases: carnitine acylcarnitine translocase (CACT), CPT1, and CPT2 (Adeva-Andany et al., 2019). CPT1 at the outer membrane of mitochondria catalyses the conversion of long-chain fatty acids acly-CoA along with carnitine and their entries into mitochondria. The transesterified acylcarnitine is transported into the mitochondrial matrix by CACT that also exports by-products of fatty acid oxidation from mitochondria to cytosol. In the mitochondrial matrix, long-chain acylcarnitine is reconverted to the respective long-chain acyl-CoA by CPT2, which is next available for β-oxidation. Three different CPT1 isozymes (CPT1A, CPT1B and CPT1C) are identified. The studies of metabolism have revealed that CPT1 may promote cancer cell proliferation and survival. miR-124-3p directly targets CPT1A, while miR-129-5p directly targets CACT in malignant prostate cells (Valentino et al., 2017). CPT1A is the target gene of miR-328-3p in breast cancer, which promotes metastasis of cancer cells by influencing fatty acid metabolism and regulating cell stemness (Zeng et al., 2022). CPT1C is identified as an miR-377-3p target in HCC cells (Zhang et al., 2022). Cholesterol homeostasis is critical for maintaining cellular function and is regulated by de novo synthesis, uptake, storage, and efflux (Goldstein & Brown, 2009). Cholesterol can be synthesised de novo by cells or from internalized low-density lipoprotein (LDL). LDL binds to the LDL receptor (LDLR) embedded in the cell membrane and is internalised into lysosomes where free cholesterol is released (Feingold et al., 2000). miR-92b suppresses LDLR expression and enhancing cervical carcinoma progression (Sun et al., 2019a). VLDLR (very-low-density lipoprotein receptor), a member of the LDLR superfamily, is also considered to play a critical role in fatty acid metabolism. Decreased VLDLR expression is inversely correlated with miR-200c expression may be involved in the development of CRC (Kim et al., 2017). Excess cholesterol is exported by ATP-binding cassette subfamily A member 1 (ABCA1) or the homodimer of ATP-binding cassette subfamily G member 1 (ABCG1), or ABCG5/8 heterodimer (Luo et al., 2020). Numerous of miRNAs have been reported to target a rather long 3’UTR of ABCA1. miR-96 regulates the expression of ABCA1 in breast cancer cell lines (Moazzeni et al., 2017), miR-101 targets ABCA1 in hepatoblastoma cells (Zhang et al., 2015c), miR-200b-3p targets ABCA1 in lung adenocarcinoma cells (Liu et al., 2019b), and miR-183 targets ABCA1 in colon cancer cells (Bi et al., 2016). miR-33a/b, encoded within the intronic region of SREBPs targets and inhibits expression of ABC transporters, ABCA1 and ABCG1, thus affecting cholesterol efflux and HDL (high-density lipoprotein) biogenesis (Marquart et al., 2010; Rayner et al., 2010). As well, ABCG1 is targeted by miR-581 in cholangiocarcinoma cells (Xu et al., 2020b). Many of enzymes involve in fatty acid metabolism are coordinately regulated by transcription factors, such as upstream stimulatory factors (USFs), SREBPs, liver X receptors (LXRs) and farnesoid X receptors (FXRs) at the transcriptional level (Calkin & Tontonoz, 2012; Wang et al., 2015). LXRs and FXRs are also regulated by cancer-associated miRNAs and activated by oxysterols and bile acids, respectively. miR-1 and miR-206 suppress lipogenesis by inhibiting LXRα and its downstream genes, SREBP1, FASN, and ACC1 (Zhong et al., 2013). miR-1247-3p targets LXRα, regulating the expression of ABCA1 in OSCC cells (Zheng et al., 2020). FXR is targeted by miR-421 in biliary tract cancer and HCC cells (Zhang et al., 2012b; Zhong et al., 2012) by miR-92 in GC cells (Duan & Fang, 2014) and by miR-382-5p in liver cancer cells (Nie et al., 2021). USFs and SREBPs play crucial roles in lipogenesis through regulated a series of genes related to fatty acid metabolism, such as FASN, ACC, ACLY, and SCD. USF1 is targeted by miR-210-3p in lung cancer cells (Chen et al., 2021b), by miR-296-5p in cholangiocarcinoma (Li et al., 2019b), and by miR‐708‐5p in GC cells (Sun et al., 2019b). USF2 is targeted by miR-362-3p in CRC cells (Christensen et al., 2013), by miR-875-5p in GC cells (Gao et al., 2022), and by miR-10a in chronic myeloid leukemia (CML) cells (Agirre et al., 2008). There are three isoforms of SREBP expressed in mammalian cells, SREBP-1a, SREBP-1c* and SREBP-2, which are transcribed by two genes, SREBF1 and SREBF2 (Goldstein & Brown, 2015). SREBP-1c is primarily responsible for the expression of lipogenic genes, although there is some functional overlap between different SREBPs, which are highly expressed in a number of tumours, such as prostate, breast cancers, HCC, and pancreatic cancer. SREBP1 is targeted by miR-18a-5p in breast cancer cells (Zhang et al., 2019b), by miR-29 in GBM cells (Ru et al., 2016), by miR-142-5p in esophageal carcer cells (Huang et al., 2019), by miR-24-3p in HCC cells (Yu et al., 2020), and by miR-132-3p in prostate cancer cells (Guo et al., 2021). miR-195 suppress the proliferation, migration, invasion, and lipogenesis of pancreatic cancer cells through regulating the expression of SREBP2 (Yu et al., 2019). PPARs are another NR family members, including three members (PPARα, PPARβ/δ, and PPARγ), that function as transcription factors to regulate the expression of genes involved in metabolic pathway (Poulsen et al., 2012). PPARα, a major isoform of PPARs, is mainly expressed in the liver, where it activates fatty acid catabolism. PPARα expression is regulated by miR-18a-3p and miR-9-5p in HCC cells (Cai et al., 2019; Luo et al., 2022), and by miR-506 in colon cancer cells (Tong et al., 2011). PPARβ/δ is expressed ubiquitously and is implicated in fatty acid oxidation, while PPARγ is mainly expressed in adipose tissue, gut and immune cells and is implicated in lipid storage and glucose metabolism. PPARγ is targeted by miR-27b-3p in thyroid cancer cells (Xu et al., 2018c), by miR-10b in esophageal cancer cells (Wu et al., 2020b), and by miR-130b in lung cancer cells (Tian et al., 2016). Because of their roles between metabolism and cellular processes, PPARs have received more interest as potential therapeutic targets for a variety of malignancies, including solid tumours, such as HCC, and CRC. In addition to the reprogramming of glucose and fatty acid metabolism, aberrant amino acid metabolism (Fig. 2) also contributes to the rapid growth and proliferation of cancer cells. Several amino acids are readily synthesized by cancer cells, and among them, serine and glycine are consumed in large numbers and are required to provide one carbon unit for downstream biosynthetic reactions (Locasale, 2013). Regulatory Roles of miRNAs Metabolic Pathways of amino acid in Cancer. This schematic diagram briefly summarizes the amino acid metabolism in cancer. Serine can be synthesized de novo in the cytosol via the serine synthesis pathway (SSP). The SSP uses the glycolytic intermediate 3PG and involves three sequential reactions catalysed by PHGDH, PAST1, and PSPH. Serine metabolism is directly linked to the folate cycle, which transfers one carbon unit between metabolites using THF as a carrier. The SHMT reaction transfers a one-carbon unit from serine to THF, generating glycine and me-THF. The glycine cleavage system (of which GLDC is a major component) also contributes to the production of me-THF in mitochondrion. This intermediate is oxidized by MTHFD1 (in cytosol) or MTHFD2 (in mitochondrion) to yield F-THF. In mitochondrion, the one-carbon unit in F-THF can be converted into formate in an MTHFD1L‑mediated reaction that regenerates ATP from ADP. Formate can be transported to the cytosol and used as substrate for the trifunctional MTHFD1, which produces F-THF for de novo purine synthesis (via GART) and me-THF for thymidylate synthesis (via TYMS). In the mitochondrion, glutamine is converted to glutamate by GLS, with the amide nitrogen released as ammonia. Glutamate can be further converted to α-KG through deamination by GLUD or transamination by glutamate-linked transaminase (e.g., GOT1/2, GPT1/2, and PSAT1) as an anaplerotic substrate in the TCA cycle for energy production. Cytosolic and mitochondrial glutamate can exchange via the aspartate/glutamate carrier. In the cytosol, glutamate together with glycine and cysteine form the GSH in two-step reaction catalysed by GCL and GSS. ASNS can transfer an amide group from glutamine to asparate to generate asparagine. For the de novo purine synthesis, PRPP, generated from the PPP, is converted into 5-PRA with the addition of an amide group from glutamine by PPAT. Glutamine can be transferred to FGAR, which in turn forms FGAM by PFAS. In the de novo purine synthesis, IMP is at branch point which the pathway diverges to the synthesis of either guanine or adenine nucleotides. Glutamine can also be transferred to XMP, which in turn forms GMP by GMPS. Furthermore, aspartate can be directly incorporated in the biosynthesis of purine (via PAICS or ADSS). Glutamate can also be directly incorporated in the proline synthesis pathway. Proline can be synthesized by conversion of glutamate to P5C by P5CS and subsequently to proline by PYCR1. Conversely, proline catabolism involves the conversion of proline to glutamine by POX, also known as PRODH, and P5CDH. Another key fate of glutamine is used for pyrimidine synthesis (via CAD and CTPS). BCAAs are reversibly metabolized by BCAT1/2, followed by irreversible decarboxylation of BCKAs by BCKDH. The end products of BCAA catabolism, acetyl-CoA and succinyl-CoA, were shown to contribute to the intermediates of the TCA cycle. Metabolic enzymes catalyzing important steps in the metabolism of these amino acids are denoted by ovals in pink, orange or blue, respectively. G6P, glucose-6-phosphate; 3PG, 3-phosphoglycerate; 3PHP, 3-phosphohydroxypyruvate; 3PS, 3-phosphoserine; α-KG, α-ketoglutarate; THF, tetrahydrofolate; me-THF, 5,10-methylene-tetrahydrofolate; F-THF, 10-formyl-tetrahydrofolate; dTMP, thymidylate; Gln, glutamine, Glu, glutamate; Ser, serine; Gly, glycine; Asp, aspartate; Asn, Asparagine; Cys, cysteine; Pro, proline; Leu, leucine; Ile, isoleucine; Val, valine; PRPP, phosphoribosyl pyrophosphate; 5-PRA, 5-phosphoribosyl-β-amine; FGAR, formylglycinamide ribonucleotide; FGAM, formylglycinamidine ribonucleotide; AIR, aminoimidazole ribonucleotide; SAICAR, succinylcarboxamide-5-aminoimidazole ribonucleotide; IMP, inosine monophosphate; AMP, adenosine monophosphate; XMP, xanthine monophosphate; GMP, guanosine monophosphate; GSA, glutamic-γ-semialdehyde; P5C, delta-1-pyrroline-5-carboxylate; GSH, glutathione; DHO, dihydroorotate; UTP, uridine triphosphate; CTP, cytidine triphosphate; BCAAs, branched-chain amino acids; BCKAs, branched-chain α-keto acids; Pathway enzymes are: PHGDH, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; SHMT1/2, serine hydroxymethyltransferase 1/2; TYMS, thymidylate synthase; MTHFD1, methylenetetrahydrofolate dehydrogenases 1; GLDC, glycine decarboxylase; GLS, glutaminase; GLUD, glutamate dehydrogenase; GPT1/2, glutamate-pyruvate transaminase 1/2; GOT1/2, glutamate–oxaloacetate transaminase 1/2; ASNS, asparagine synthetase; GCL, glutamate cysteine ligase; GSS, glutathione synthetase; P5CS, P5C synthase; P5CDH, P5C dehydrogenase; PYCR1, P5C reductase 1; PRODH/POX, proline dehydrogenase/proline oxidase; BCAT1/2, branched-chain amino acid transaminase 1/2; BCKDH, BCKA dehydrogenase enzyme complex; CAD, carbamoylphosphate synthetase, including carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydrooratase; CTPS, CTP synthase; PPAT, phosphoribosyl pyrophosphate amidotransferase; GART, phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase; PFAS, phosphoribosylformylglycinamidine synthase; PAICS, phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase; ADSS, adenylosuccinate synthase; GMPS, guanine monophosphate synthase Phosphoglycerate dehydrogenase (PHGDH) functions at the branching point of glycolysis-dependent serine and glycine metabolism (Achouri et al., 1997). Phosphoserine aminotransferase 1 (PSAT1) then convers 3-phosphohydroxypyruvate into 3-phosphoserine through a glutamate-dependent transamination reaction. 3-phosphoserine is dephosphorylated by the final enzyme in the pathway, phosphoserine phosphatase (PSPH), to produce serine (Snell, 1984). PHGDH, a promising therapeutic target for cancer, is modulated by miR-107/103a-3p in HCC cells (Zhang et al., 2021b), and by miR-940 in CRC cells (Table 1) (Sun et al., 2021). PSAT1 is responsible for miRNA-195-5p-reduced cisplatin resistance and angiogenesis in ovarian cancer (Dai et al., 2019), for miR-340- and miR-365-suppressed cell growth, and invasion in esophageal squamous cell carcinoma (ESCC) (Sun et al., 2018; Yan et al., 2015), and for miR-424-induced apoptosis in CRC (Fang et al., 2018). Until now, no major cancer-associated miRNAs are known to directly regulate PSPH. Subsequent to its production, serine can be converted to glycine by serine hydroxymethyltransferase 1 (SHMT1) (Stover & Schirch, 1990), which is regulated by miR-218-5p and miR-198 in lung adenocarcinoma cells (Wu et al., 2016a; Yang et al., 2019). SHMT2, the isoform enzyme located in mitochondrion, performs the same reaction like SHMT1. SHMT2 is regulated by miR-615-5p in HCC cells (Wu et al., 2016b), and by miR-193b in breast cancer cells (Leivonen et al., 2011). Mitochondrially localized glycine can be further consumed by the glycine cleavage system (GCS), a multiprotein complex that includes glycine decarboxylase (GLDC) (Kikuchi et al., 2008). GLDC is downregulated by miR-30d-5p in HCC cells (Zhuang et al., 2019). One carbon pool is managed principally by methylenetetrahydrofolate dehydrogenases (MTHFD1, 1L, and 2) and supports a variety of cellular methylation reactions through the production of S-adenosyl methionine (SAM), as well as nucleotide biosynthesis via thymidylate synthase (TYMS) and the trifunctional enzyme GART (phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase) (Tibbetts & Appling, 2010). miR-338-3p regulates the proliferation, migration, and invasion in arbutin-treated osteosarcoma cells via directly targeting MTHFD1L (Wang et al., 2021b). MTHFD2, a mitochondrial enzyme involved in one carbon metabolism, is directly targeted by miR-9 and miR-22 in breast cancer cells (Koufaris et al., 2016; Selcuklu et al., 2012), by miR-22 in GC cells (Tong et al., 2020), by miR-92a and miR-504-3p in acute myeloid leukemia (AML) cells (Gu et al., 2017; Li et al., 2019c), by miR-940 in GBM cells (Xu et al., 2019), by miR-30a-3p in lung adenocarcinoma cells (Shi et al., 2021), and by miR-33a-5p in CRC cells (Yan et al., 2019). No major cancer-associated miRNAs involved in MTHFD1 and GART regulation have been identified. The TYMS gene is regulated by miR-192/215, miR-197, miR-330, and miR-375-3p in CRC cells (Boni et al., 2010; Sun et al., 2015; Xu et al., 2017; Xu et al., 2020c), by miR-140-3p in lung adenocarcinoma cells (Wan et al., 2021), and by miR-433 in ovarian cancer cells (Gotanda et al., 2013), impacting the resistance to 5-fluorouracil. Apart from glucose, glutamine also participates in multiple biological processes required for cancer cell growth and proliferation (DeBerardinis et al., 2007). Glutamine can replenish the TCA cycle as an anaplerotic substrate and is an indispensable nitrogen donor in the biosynthesis of purines, pyrimidines, non-essential amino acids, NAD, and glucosamine. Although glutamine can be generated biosynthetically, most cancer cell lines exclusively convert glutamine to glutamate through the actions of phosphoribosyl pyrophosphate amidotransferase (PPAT), phosphoribosylformylglycinamidine synthase (PFAS), CTP synthase 1/2 (CTPS1/2), CAD (carbamoylphosphate synthetase, including carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), and guanine monophosphate synthase (GMPS). So far, no revelant studies have focused on miRNAs that target these genes in cancer cells. Glutaminase (GLS) catalyses the hydrolysis of glutamine to glutamate and ammonia. Study has found that miR-23, which directly targets GLS, enhances glutamine catabolism through increased mitochondrial GLS expression (Gao et al., 2009). GLS is also directly targeted by miR-122 in HCC cells (Sengupta et al., 2020), by miR-141-3p in osteosarcoma cells (Zhou, 2021), by miR-137 in colon cancer and melanoma cells (Fan et al., 2022; Luan et al., 2018), and by miR-203 in melanoma cells (Chang et al., 2017). Glutamate maintains the viability and proliferation of cancer cells by replenishing the TCA cycle intermediate, α-KG, mediated by either glutamate dehydrogenase (GLUD), alanine aminotransferase (also known as glutamate-pyruvate transaminase, GPT), or aspartate aminotransferase (also known as glutamate–oxaloacetate transaminase 1/2, GOT1/2) (Altman et al., 2016). Some studies have reported that GOT1 is directly targeted by miR-9 in pancreatic cancer and melanoma cells (Wang et al., 2019a; Zhang et al., 2018), by miR-377-3p in NSCLC cells (Zhu et al., 2020), by miR-429 in HCC cells (Liu & Shen, 2020), and by miR-433-3p in pancreatic cancer cells (Zhou et al., 2021). GOT2, one isoform enzyme located in mitochondrion, is regulated by miR-1298-5p and miR-520a-5p in NSCLC cells (Guan et al., 2021; Jin et al., 2020). Proline plays a special role in cancer metabolism that can be a source of ATP as well as a supply of α-KG for anaplerosis. Glutamate can be converted to proline through delta-1-pyrroline-5-carboxylate, catalysed by delta-1-pyrroline-5-carboxylate synthetase (P5CS) and P5C reductase 1 (PYCR1). Conversely, proline catabolism involves the conversion of proline to glutamine by proline oxidase (POX), also known as proline dehydrogenase (PRODH), and P5C dehydrogenase (P5CDH) (Liu et al., 2012). Few studies have focused on miRNAs that target these enzymes in cancer cells. Studies have found that PYCR1 expression is regulated by miR-328-3p in lung adenocarcinoma cells (Lu et al., 2021), by miR-1207-5p in prostate cancer cells (Xu et al., 2021b), and by miR-488 in NSCLC cells (Wang et al., 2019b), while PRODH/POX expression is regulated by miR-23b* in RCC cells (Liu et al., 2010). Glutamine can also participate in the de novo synthesis of GSH by providing glutamate. Intracellular GSH is synthesised by glutamate cysteine ligase (GCL) and glutathione synthetase (GSS) in a two-step reaction from glutamate, cysteine, and glycine, the latter being derived mainly from serine (Bansal & Simon, 2018). GCL is a heterodimer consisting of a catalytic subunit (GCLC) and a regulatory subunit (GCLM). There is little experimental-based evidence of cancer-associated miRNAs that regulate these enzymes. However, GCLC expression is attenuated by miR-18a in myc-driven liver tumours that are highly susceptible to exogenous oxidative stress (Anderton et al., 2017). Asparagine is moderately abundant in circulation, and unlike other amino acids, is only used for protein biosynthesis. Asparagine synthetase (ASNS) levels are correlated with tumour aggressiveness, potentially through a mechanism in which asparagine suppresses apoptosis (Zhang et al., 2014). However, there are no major cancer-associated miRNAs involved in ASNS regulation. A key fate of aspartate is its assimilation into nucleotide precursors in the de novo biosynthesis of pyrimidine via CAD and the contribution of nitrogen to the de novo purine biosynthetic pathway via PAICS (phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase) or ADSS (adenylosuccinate synthase) (Tsun & Possemato, 2015). PAICS, an oncogenic factor in various cancers, is regulated by miR-504 in cervical cancer cells (Qu et al., 2020b), by miR-128 in CRC and bladder cancer cells (Agarwal et al., 2020; Chakravarthi et al., 2018), by miR-142-5p in NSCLC cells (Tang et al., 2021), and by miR-4731-5p in breast cancer cells (Lang et al., 2022), thus modulating EMT (epithelial-mesenchymal transition). Branched-chain amino acids (BCAAs), including leucine, isoleucine, and valine, are essential amino acids for mammals. BCAA catabolism is mediated by cytosolic branched-chain amino acid transaminase 1 (BCAT1) and mitochondrial BCAT2, which transfer the amino groups from BCAAs to α-KG to produce branched-chain α-keto acids (BCKAs) and glutamate (Sivanand & Vander Heiden, 2020). In both enzymes, BCAT1 is the major isoform implicated in cancer growth and regulated by multiple factors, including miRNAs. Studies show that miR-218 inhibits tumour growth and increases chemosensitivity by targeting BCAT1 in prostate cancer cells (Zhu et al., 2017). BCAT1 is also targeted by miR-98-5p in GC and CRC cells (Ye et al., 2022; Zhan et al., 2021), by miR-124-3p in ESCC cells (Zeng et al., 2019), by miR-506 in glioma cells (Ye et al., 2020b), and by miR-934 in osteosarcoma cells (Wei et al., 2020). BCKAs are then catabolized by BCKA dehydrogenase enzyme complex (BCKDH) in the mitochondria, which eventually metabolized into acetyl-CoA and succinyl-CoA that can feed into the TCA cycle and can contribute to energy production. Altered metabolism is considered a core hallmark of cancer. Precise regulation of metabolic enzymes promotes metabolic reprogramming in cancer cells, resulting in enhanced nutrient uptake to supply energy and biosynthetic pathways required for malignant progression. miRNAs are noncoding RNAs that have been considered as master regulator roles of gene expression. miRNAs participate in cancer metabolism not only by directly regulate key enzymes or transporter proteins that involved in metabolism, but also by regulate transcription factors such as NRs, which in turn regulate metabolic enzymes. The role of miRNAs in cancer metabolism is a rapidly expanding research area. miRNAs participate in the control of glucose, fatty acid, and amino acid metabolic reprogramming, thus affecting cancer progression. miRNA mimics or inhibitors can be used to modulate the activity of miRNAs. Use of miRNA mimics or antisense oligonucleotides will lead to a new therapeutic strategy for tumours. Several clinical trials investigating the role of miRNA-based therapy for cancer have been launched that may lead to novel therapeutic interventions in the future. For example, miR-34a is described as a master tumour suppressor in many types of cancers, such as breast cancer, CRC, and lung cancer. miR-34a, which plays a role in the regulation of glycolysis (HK2, GPI, and LDHA), and the TCA cycle (PDK1), has been proved to affect almost the whole cancer progression process. MRX34, a liposomal miR-34a mimic, was already in phase 1 clinical trial in 2014 (NCT01829971) for the management of multiple solid tumours. However, the challenges of miRNA-based therapies are to find the best miRNA for a particular cancer type, suitable delivery system, and off-target effects. Since these challenges, molecular strategies of miRNA-based cancer therapy are still in infancy. More research is clearly warranted in this area. It is undeniable that the studies of miRNAs in metabolic reprogramming have brought a new dawn for cancer therapy. The authors confirm that the data supporting the findings of this study are available within the article. Achouri, Y., et al. (1997). Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. The Biochemical Journal, 323(Pt 2), 365–370. Adeva-Andany, M. M., et al. (2019). Mitochondrial beta-oxidation of saturated fatty acids in humans. Mitochondrion, 46, 73–90. Agarwal, S., et al. (2020). PAICS, a purine nucleotide metabolic enzyme, is involved in tumor growth and the metastasis of colorectal cancer. Cancers (basel), 12(4), 772. Agirre, X., et al. (2008). Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Molecular Cancer Research, 6(12), 1830–1840. Akram, M. (2014). Citric acid cycle and role of its intermediates in metabolism. Cell Biochemistry and Biophysics, 68(3), 475–478. Altman, B. J., Stine, Z. E., & Dang, C. V. (2016). From Krebs to clinic: Glutamine metabolism to cancer therapy. Nature Reviews Cancer, 16(10), 619–634. Anderson, N. M., et al. (2018). The emerging role and targetability of the TCA cycle in cancer metabolism. Protein & Cell, 9(2), 216–237. Anderton, B., et al. (2017). MYC-driven inhibition of the glutamate-cysteine ligase promotes glutathione depletion in liver cancer. EMBO Reports, 18(4), 569–585. Bai, Y., et al. (2015). X-ray structure of a mammalian stearoyl-CoA desaturase. Nature, 524(7564), 252–256. Bansal, A., & Simon, M. C. (2018). Glutathione metabolism in cancer progression and treatment resistance. Journal of Cell Biology, 217(7), 2291–2298. Barajas, J. M., et al. (2018). The role of miR-122 in the dysregulation of glucose-6-phosphate dehydrogenase (G6PD) expression in hepatocellular cancer. Science and Reports, 8(1), 9105. Benjamin, D. I., Cravatt, B. F., & Nomura, D. K. (2012). Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metabolism, 16(5), 565–577. Bi, D. P., et al. (2016). MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncology Reports, 35(5), 2873–2879. Bienertova-Vasku, J., Sana, J., & Slaby, O. (2013). The role of microRNAs in mitochondria in cancer. Cancer Letters, 336(1), 1–7. Boguslawska, J., et al. (2019). MicroRNA-mediated metabolic reprograming in renal cancer. Cancers (basel), 11(12), 1825. Boni, V., et al. (2010). miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Molecular Cancer Therapeutics, 9(8), 2265–2275. Bracken, C. P., Scott, H. S., & Goodall, G. J. (2016). A network-biology perspective of microRNA function and dysfunction in cancer. Nature Reviews Genetics, 17(12), 719–732. Bricker, D. K., et al. (2012). A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science, 337(6090), 96–100. Butler, L. M., et al. (2020). Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Advanced Drug Delivery Reviews, 159, 245–293. Cai, K., et al. (2019). Long non-coding RNA LINC00467 regulates hepatocellular carcinoma progression by modulating miR-9-5p/PPARA expression. Open Biology, 9(9), 190074. Calkin, A. C., & Tontonoz, P. (2012). Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nature Reviews Molecular Cell Biology, 13(4), 213–224. Cannistraci, A., et al. (2022). MiR-378a inhibits glucose metabolism by suppressing GLUT1 in prostate cancer. Oncogene. https://doi.org/10.1038/s41388-022-02178-0 Cao, H., et al. (2017). Curcumin inhibits prostate cancer by targeting PGK1 in the FOXD3/miR-143 axis. Cancer Chemotherapy and Pharmacology, 79(5), 985–994. Chai, S., et al. (2016). Octamer 4/microRNA-1246 signaling axis drives Wnt/beta-catenin activation in liver cancer stem cells. Hepatology, 64(6), 2062–2076. Chakravarthi, B., et al. (2018). A role for de novo purine metabolic enzyme PAICS in bladder cancer progression. Neoplasia, 20(9), 894–904. Chang, X., et al. (2017). Sensitization of melanoma cells to temozolomide by overexpression of microRNA 203 through direct targeting of glutaminase-mediated glutamine metabolism. Clinical and Experimental Dermatology, 42(6), 614–621. Chen, B., et al. (2015). miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Letters, 356(2 Pt B), 410–417. Chen, H., Gao, S., & Cheng, C. (2018b). MiR-323a-3p suppressed the glycolysis of osteosarcoma via targeting LDHA. Human Cell, 31(4), 300–309. Chen, J., et al. (2018a). MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Proliferation, 51(6), e12510. Chen, J., et al. (2019a). Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Molecular Cancer, 18(1), 33. Chen, L., et al. (2021a). Quercetin suppresses cell survival and invasion in oral squamous cell carcinoma via the miR-1254/CD36 cascade in vitro. Human and Experimental Toxicology, 40(9), 1413–1421. Chen, Q., et al. (2021b). miR-210-3p promotes lung cancer development and progression by modulating USF1 and PCGF3. Oncotargets and Therapy, 14, 3687–3700. Chen, R., Zhang, Y., & Zhang, X. (2019c). MiR-1254 functions as a tumor suppressor in oral squamous cell carcinoma by targeting CD36. Technology in Cancer Research & Treatment, 18, 1533033819859447. Chen, Z., et al. (2019b). MicroRNA-450b-3p inhibits cell growth by targeting phosphoglycerate kinase 1 in hepatocellular carcinoma. Journal of Cellular Biochemistry, 120(11), 18805–18815. Christensen, L. L., et al. (2013). MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. International Journal of Cancer, 133(1), 67–78. Coda, D. M., et al. (2015). SMYD1 and G6PD modulation are critical events for miR-206-mediated differentiation of rhabdomyosarcoma. Cell Cycle, 14(9), 1389–1402. Cohen, P. (1999). The Croonian Lecture 1998. Identification of a protein kinase cascade of major importance in insulin signal transduction. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 354(1382), 485–495. Convertini, P., et al. (2016). The contribution of the citrate pathway to oxidative stress in Down syndrome. Immunology, 149(4), 423–431. Croce, C. M. (2008). Oncogenes and cancer. New England Journal of Medicine, 358(5), 502–511. Cruz-Gil, S., et al. (2018). Targeting the lipid metabolic axis ACSL/SCD in colorectal cancer progression by therapeutic miRNAs: MiR-19b-1 role. Journal of Lipid Research, 59(1), 14–24. Cui, J., et al. (2018). MicroRNA-206 suppresses proliferation and predicts poor prognosis of HR-HPV-positive cervical cancer cells by targeting G6PD. Oncology Letters, 16(5), 5946–5952. Cui, K., et al. (2019). Long noncoding RNA DIO3OS interacts with miR-122 to promote proliferation and invasion of pancreatic cancer cells through upregulating ALDOA. Cancer Cell International, 19, 202. Currie, E., et al. (2013). Cellular fatty acid metabolism and cancer. Cell Metabolism, 18(2), 153–161. Dai, D. W., et al. (2013). Decreased miR-106a inhibits glioma cell glucose uptake and proliferation by targeting SLC2A3 in GBM. BMC Cancer, 13, 478. Dai, J., et al. (2019). Overexpression of microRNA-195-5p reduces cisplatin resistance and angiogenesis in ovarian cancer by inhibiting the PSAT1-dependent GSK3beta/beta-catenin signaling pathway. Journal of Translational Medicine, 17(1), 190. DeBerardinis, R. J., et al. (2007). Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A, 104(49), 19345–19350. Deng, Y. H., et al. (2018). MicroRNA-23a promotes colorectal cancer cell survival by targeting PDK4. Experimental Cell Research, 373(1–2), 171–179. Ding, X., et al. (2017). miR-148b inhibits glycolysis in gastric cancer through targeting SLC2A1. Cancer Medicine, 6(6), 1301–1310. Duan, J. H., & Fang, L. (2014). MicroRNA-92 promotes gastric cancer cell proliferation and invasion through targeting FXR. Tumour Biology, 35(11), 11013–11019. Ebron, J. S., et al. (2019). MiR-644a disrupts oncogenic transformation and warburg effect by direct modulation of multiple genes of tumor-promoting pathways. Cancer Research, 79(8), 1844–1856. Fan, D., et al. (2016b). Upregulation of miR-501-5p activates the wnt/beta-catenin signaling pathway and enhances stem cell-like phenotype in gastric cancer. Journal of Experimental & Clinical Cancer Research, 35(1), 177. Fan, J. Y., et al. (2016a). MicroRNA-144 mediates metabolic shift in ovarian cancer cells by directly targeting Glut1. Tumour Biology, 37(5), 6855–6860. Fan, W. H., et al. (2022). Curcumin synergizes with cisplatin to inhibit colon cancer through targeting the MicroRNA-137-glutaminase axis. Current Medical Science, 42(1), 108–117. Fang, R., et al. (2012). MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. Journal of Biological Chemistry, 287(27), 23227–23235. Fang, Y., et al. (2018). miR-424 targets AKT3 and PSAT1 and has a tumor-suppressive role in human colorectal cancer. Cancer Management and Research, 10, 6537–6547. Fei, X., et al. (2012). MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Letters, 586(4), 392–397. Feingold, K.R. (2000). Introduction to lipids and lipoproteins, In K. R. Feingold, et al., (Eds.), Endotext Feng, L., et al. (2019). miR-497–5p inhibits gastric cancer cell proliferation and growth through targeting PDK3. Bioscience Reports. https://doi.org/10.1042/BSR20190654 Gaal, Z. (2021). MicroRNAs and metabolism: Revisiting the warburg effect with emphasis on epigenetic background and clinical applications. Biomolecules, 11(10), 1531. Gao, P., et al. (2009). c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature, 458(7239), 762–765. Gao, S., et al. (2022). hsa-miR-875-5p inhibits tumorigenesis and suppresses TGF-beta signalling by targeting USF2 in gastric cancer. Journal of Translational Medicine, 20(1), 115. Ge, J., et al. (2019). miR-548c-5p inhibits colorectal cancer cell proliferation by targeting PGK1. Journal of Cellular Physiology, 234(10), 18872–18878. Gebert, L. F. R., & MacRae, I. J. (2019). Regulation of microRNA function in animals. Nature Reviews Molecular Cell Biology, 20(1), 21–37. Gibbings, D., et al. (2012). Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nature Cell Biology, 14(12), 1314–1321. Goldstein, J. L., & Brown, M. S. (2009). The LDL receptor. Arteriosclerosis, Thrombosis, and Vascular Biology, 29(4), 431–438. Goldstein, J. L., & Brown, M. S. (2015). A century of cholesterol and coronaries: From plaques to genes to statins. Cell, 161(1), 161–172. Goodall, G. J., & Wickramasinghe, V. O. (2021). RNA in cancer. Nature Reviews Cancer, 21(1), 22–36. Gorbea, C., Mosbruger, T., & Cazalla, D. (2017). A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis. Nature, 550(7675), 275–279. Gotanda, K., et al. (2013). MicroRNA-433 negatively regulates the expression of thymidylate synthase (TYMS) responsible for 5-fluorouracil sensitivity in HeLa cells. BMC Cancer, 13, 369. Gray, L. R., Tompkins, S. C., & Taylor, E. B. (2014). Regulation of pyruvate metabolism and human disease. Cellular and Molecular Life Sciences, 71(14), 2577–2604. Gregersen, L. H., et al. (2012). MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer, 12, 232. Gu, Y., et al. (2017). miR-92a inhibits proliferation and induces apoptosis by regulating methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) expression in acute myeloid leukemia. Oncology Research, 25(7), 1069–1079. Gu, Y., et al. (2020). Loss of miR-192-5p initiates a hyperglycolysis and stemness positive feedback in hepatocellular carcinoma. Journal of Experimental & Clinical Cancer Research, 39(1), 268. Guan, H., et al. (2021). Circular RNA circ_0003028 contributes to tumorigenesis by regulating GOT2 via miR-1298-5p in non-small cell lung cancer. Bioengineered, 12(1), 2326–2340. Guo, J., et al. (2019). MiR-204-3p inhibited the proliferation of bladder cancer cells via modulating lactate dehydrogenase-mediated glycolysis. Frontiers in Oncology, 9, 1242. Guo, Q., et al. (2020). MicroRNA-206 inhibits tumor metastasis of nasopharyngeal carcinoma through targeting G6PD. Journal of Biological Regulators and Homeostatic Agents. https://doi.org/10.23812/20-36A Guo, S., et al. (2021). LncRNA PCA3 promotes antimony-induced lipid metabolic disorder in prostate cancer by targeting MIR-132-3 P/SREBP1 signaling. Toxicology Letters, 348, 50–58. Guo, W., et al. (2015). MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology, 62(4), 1132–1144. Ha, M., & Kim, V. N. (2014). Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology, 15(8), 509–524. Han, L., et al. (2016). MicroRNA-497 downregulation contributes to cell proliferation, migration, and invasion of estrogen receptor alpha negative breast cancer by targeting estrogen-related receptor alpha. Tumour Biology, 37(10), 13205–13214. Han, R. L., et al. (2017). miR-383 inhibits ovarian cancer cell proliferation, invasion and aerobic glycolysis by targeting LDHA. Neoplasma, 64(2), 244–252. Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discovery, 12(1), 31–46. Hanahan, D., & Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell, 144(5), 646–674. Hatziapostolou, M., et al. (2011). An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell, 147(6), 1233–1247. He, Y., et al. (2018a). LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Molecular Carcinogenesis, 57(6), 772–783. He, Y., et al. (2019). Analysis of miRNA-mRNA network reveals miR-140-5p as a suppressor of breast cancer glycolysis via targeting GLUT1. Epigenomics, 11(9), 1021–1036. He, Z., et al. (2018b). MiR-422a regulates cellular metabolism and malignancy by targeting pyruvate dehydrogenase kinase 2 in gastric cancer. Cell Death & Disease, 9(5), 505. Hou, L., et al. (2019). Interfering cellular lactate homeostasis overcomes Taxol resistance of breast cancer cells through the microRNA-124-mediated lactate transporter (MCT1) inhibition. Cancer Cell International, 19, 193. Hua, S., et al. (2018). miR-142-3p inhibits aerobic glycolysis and cell proliferation in hepatocellular carcinoma via targeting LDHA. Biochemical and Biophysical Research Communications, 496(3), 947–954. Huang, C. M., et al. (2019). Disruption of cancer metabolic SREBP1/miR-142–5p suppresses epithelial-mesenchymal transition and stemness in esophageal carcinoma. Cells, 9(1), 7. Huang, J., et al. (2016). c-Myc modulates glucose metabolism via regulation of miR-184/PKM2 pathway in clear-cell renal cell carcinoma. International Journal of Oncology, 49(4), 1569–1575. Jauhari, A., Singh, T., & Yadav, S. (2022). Neurodevelopmental disorders and neurotoxicity: MicroRNA in focus. Journal of Chemical Neuroanatomy, 120, 102072. Jiang, N., et al. (2020). HIF-1a-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/beta-catenin and Notch signaling. Theranostics, 10(6), 2553–2570. Jiang, S., et al. (2012). A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO Journal, 31(8), 1985–1998. Jin, M., et al. (2020). High circ-SEC31A expression predicts unfavorable prognoses in non-small cell lung cancer by regulating the miR-520a-5p/GOT-2 axis. Aging (albany NY), 12(11), 10381–10397. Jones, S. F., & Infante, J. R. (2015). Molecular pathways: fatty acid synthase. Clinical Cancer Research, 21(24), 5434–5438. Ju, J., et al. (2020). miR-150 regulates glucose utilization through targeting GLUT4 in insulin-resistant cardiomyocytes. Acta Biochimica Et Biophysica Sinica (shanghai), 52(10), 1111–1119. Kefas, B., et al. (2010). Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro-Oncology, 12(11), 1102–1112. Kikuchi, G., et al. (2008). Glycine cleavage system: Reaction mechanism, physiological significance, and hyperglycinemia. Proceedings of the Japan Academy. Series b, Physical and Biological Sciences, 84(7), 246–263. Kim, B. K., et al. (2017). Decreased expression of VLDLR is inversely correlated with miR-200c in human colorectal cancer. Molecular Carcinogenesis, 56(6), 1620–1629. Kim, H. R., et al. (2013). p53 regulates glucose metabolism by miR-34a. Biochemical and Biophysical Research Communications, 437(2), 225–231. Komoll, R. M., et al. (2021). MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. Journal of Hepatology, 74(1), 122–134. Koufaris, C., et al. (2016). Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene, 35(21), 2766–2776. Koundouros, N., & Poulogiannis, G. (2020). Reprogramming of fatty acid metabolism in cancer. British Journal of Cancer, 122(1), 4–22. Krokker, L., et al. (2019). Differentially expressed miRNAs influence metabolic processes in pituitary oncocytoma. Neurochemical Research, 44(10), 2360–2371. Kwak, S., et al. (2022). miR-3189-targeted GLUT3 repression by HDAC2 knockdown inhibits glioblastoma tumorigenesis through regulating glucose metabolism and proliferation. Journal of Experimental & Clinical Cancer Research, 41(1), 87. Lang, L., et al. (2022). Tumor suppressive role of microRNA-4731-5p in breast cancer through reduction of PAICS-induced FAK phosphorylation. Cell Death Discovery, 8(1), 154. Lee, R. C., Feinbaum, R. L., & Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75(5), 843–854. Leivonen, S. K., et al. (2011). Identification of miR-193b targets in breast cancer cells and systems biological analysis of their functional impact. Molecular and Cellular Proteomics, 10(7), M110 005322. Li, F., et al. (2020). PGAM1, regulated by miR-3614-5p, functions as an oncogene by activating transforming growth factor-beta (TGF-beta) signaling in the progression of non-small cell lung carcinoma. Cell Death & Disease, 11(8), 710. Li, G., et al. (2017b). The microRNA-182-PDK4 axis regulates lung tumorigenesis by modulating pyruvate dehydrogenase and lipogenesis. Oncogene, 36(7), 989–998. Li, G., Li, Y., & Wang, D. Y. (2021b). Overexpression of miR-329-3p sensitizes osteosarcoma cells to cisplatin through suppression of glucose metabolism by targeting LDHA. Cell Biology International, 45(4), 766–774. Li, H., et al. (2019a). MiR-122 promotes the development of colon cancer by targeting ALDOA in vitro. Technology in Cancer Research & Treatment, 18, 1533033819871300. Li, H., et al. (2021d). MiR-4310 regulates hepatocellular carcinoma growth and metastasis through lipid synthesis. Cancer Letters, 519, 161–171. Li, K. K., et al. (2009). miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Human Pathology, 40(9), 1234–1243. Li, K., Zhu, X., & Yuan, C. (2021a). Inhibition of miR-185-3p confers erlotinib resistance through upregulation of PFKL/MET in lung cancers. Frontiers in Cell and Developmental Biology, 9, 677860. Li, L., et al. (2017a). miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Letters, 400, 89–98. Li, L., et al. (2018). miR-449a suppresses LDHA-mediated glycolysis to enhance the sensitivity of non-small cell lung cancer cells to ionizing radiation. Oncology Research, 26(4), 547–556. Li, S. M., et al. (2019c). Upregulation of miR-504-3p is associated with favorable prognosis of acute myeloid leukemia and may serve as a tumor suppressor by targeting MTHFD2. European Review for Medical and Pharmacological Sciences, 23(3), 1203–1213. Li, T., et al. (2016). MicroRNA-224 sustains Wnt/beta-catenin signaling and promotes aggressive phenotype of colorectal cancer. Journal of Experimental & Clinical Cancer Research, 35, 21. Li, X., et al. (2021c). miR-140-5p aggravates insulin resistance via directly targeting GYS1 and PPP1CC in insulin-resistant HepG2 cells. Diabetes Metab Syndr Obes, 14, 2515–2524. Li, Z., et al. (2019b). Up-regulation of ZFAS1 indicates dismal prognosis for cholangiocarcinoma and promotes proliferation and metastasis by modulating USF1 via miR-296-5p. Journal of Cellular and Molecular Medicine, 23(12), 8258–8268. Li, Z., & Zhang, H. (2016). Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cellular and Molecular Life Sciences, 73(2), 377–392. Liang, Y., et al. (2020). Dichloroacetate restores colorectal cancer chemosensitivity through the p53/miR-149-3p/PDK2-mediated glucose metabolic pathway. Oncogene, 39(2), 469–485. Lima Queiroz, A., et al. (2018). miR-126-5p targets Malate Dehydrogenase 1 in non-small cell lung carcinomas. Biochemical and Biophysical Research Communications, 499(2), 314–320. Liu, F., et al. (2022). miR-137/ERRalpha axis mediates chemoresistance of nasopharyngeal carcinoma cells. Journal of Cell Communication and Signaling, 16(1), 103–113. Liu, J., et al. (2020b). A microRNA-messenger RNA regulatory network and its prognostic value in cervical cancer. DNA and Cell Biology, 39(7), 1328–1346. Liu, K., et al. (2019b). MiR-200b-3p functions as an oncogene by targeting ABCA1 in lung adenocarcinoma. Technology in Cancer Research & Treatment, 18, 1533033819892590. Liu, Q., et al. (2021). Glycogen accumulation and phase separation drives liver tumor initiation. Cell, 184(22), 5559-5576 e19. Liu, S., et al. (2017). miR-142-5p promotes development of colorectal cancer through targeting SDHB and facilitating generation of aerobic glycolysis. Biomedicine & Pharmacotherapy, 92, 1119–1127. Liu, S., Chen, Q., & Wang, Y. (2020a). MiR-125b-5p suppresses the bladder cancer progression via targeting HK2 and suppressing PI3K/AKT pathway. Human Cell, 33(1), 185–194. Liu, W., et al. (2010). miR-23b targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene, 29(35), 4914–4924. Liu, W., et al. (2012). Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proceedings of the National Academy of Sciences, 109(23), 8983–8988. Liu, X., & Shen, Z. (2020). LncRNA TMPO-AS1 Aggravates the Development of Hepatocellular Carcinoma via miR-429/GOT1 Axis. American Journal of the Medical Sciences, 360(6), 711–720. Liu, Y., et al. (2018). MiR-22-3p targeting alpha-enolase 1 regulates the proliferation of retinoblastoma cells. Biomedicine & Pharmacotherapy, 105, 805–812. Liu, Y., et al. (2019a). A novel oxoglutarate dehydrogenase-like mediated miR-214/TWIST1 negative feedback loop inhibits pancreatic cancer growth and metastasis. Clinical Cancer Research, 25(17), 5407–5421. Locasale, J. W. (2013). Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nature Reviews Cancer, 13(8), 572–583. Lone, S. N., et al. (2018). Triose-phosphate isomerase is a novel target of miR-22 and miR-28, with implications in tumorigenesis. Journal of Cellular Physiology, 233(11), 8919–8929. Lu, J., et al. (2019). miR-603 targeted hexokinase-2 to inhibit the malignancy of ovarian cancer cells. Archives of Biochemistry and Biophysics, 661, 1–9. Lu, J., et al. (2021). MiR-328-3p inhibits lung adenocarcinoma-genesis by downregulation PYCR1. Biochemical and Biophysical Research Communications, 550, 99–106. Lu, M., et al. (2015). MicroRNA-320a sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by targeting ARPP-19 and ERRgamma. Science and Reports, 5, 8735. Luan, W., et al. (2018). miR-137 inhibits glutamine catabolism and growth of malignant melanoma by targeting glutaminase. Biochemical and Biophysical Research Communications, 495(1), 46–52. Luo, J., Yang, H., & Song, B. L. (2020). Mechanisms and regulation of cholesterol homeostasis. Nature Reviews Molecular Cell Biology, 21(4), 225–245. Luo, Y. Y., et al. (2022). Hsa_Circ_0098181 suppresses hepatocellular carcinoma by sponging miR-18a-3p and targeting PPARA. Frontiers in Pharmacology, 13, 819735. Ma, K., et al. (2005). Cloning of the rat pyruvate dehydrogenase kinase 4 gene promoter: Activation of pyruvate dehydrogenase kinase 4 by the peroxisome proliferator-activated receptor gamma coactivator. Journal of Biological Chemistry, 280(33), 29525–29532. Ma, X., et al. (2014). Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nature Communications, 5, 5212. Macheda, M. L., Rogers, S., & Best, J. D. (2005). Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. Journal of Cellular Physiology, 202(3), 654–662. Marquart, T. J., et al. (2010). miR-33 links SREBP-2 induction to repression of sterol transporters. Proceedings of the National Academy of Sciences of the United States of America, 107(27), 12228–12232. Miao, Y., et al. (2020). MiR-5683 suppresses glycolysis and proliferation through targeting pyruvate dehydrogenase kinase 4 in gastric cancer. Cancer Medicine, 9(19), 7231–7243. Moazzeni, H., Najafi, A., & Khani, M. (2017). Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Molecular and Cellular Probes, 34, 45–52. Morimoto, A., et al. (2017). An HNF4alpha-microRNA-194/192 signaling axis maintains hepatic cell function. Journal of Biological Chemistry, 292(25), 10574–10585. Mutlu, M., et al. (2016). miR-564 acts as a dual inhibitor of PI3K and MAPK signaling networks and inhibits proliferation and invasion in breast cancer. Science and Reports, 6, 32541. Muys, B. R., et al. (2019). miR-450a acts as a tumor suppressor in ovarian cancer by regulating energy metabolism. Cancer Research, 79(13), 3294–3305. Ni, Y., et al. (2020). miR-15a-5p inhibits metastasis and lipid metabolism by suppressing histone acetylation in lung cancer. Free Radical Biology & Medicine, 161, 150–162. Nie, X., et al. (2021). miRNA-382-5p suppresses the expression of farnesoid X receptor to promote progression of liver cancer. Cancer Manag Res, 13, 8025–8035. Ning, B. F., et al. (2014). Hepatocyte nuclear factor 4alpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology, 60(5), 1607–1619. Park, Y. Y., et al. (2013). Tat-activating regulatory DNA-binding protein regulates glycolysis in hepatocellular carcinoma by regulating the platelet isoform of phosphofructokinase through microRNA 520. Hepatology, 58(1), 182–191. Pascual, G., et al. (2017). Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature, 541(7635), 41–45. Patra, K. C., & Hay, N. (2014). The pentose phosphate pathway and cancer. Trends in Biochemical Sciences, 39(8), 347–354. Pavlova, N. N., & Thompson, C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metabolism, 23(1), 27–47. Peschiaroli, A., et al. (2013). miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene, 32(6), 797–802. Poulsen, L., Siersbaek, M., & Mandrup, S. (2012). PPARs: Fatty acid sensors controlling metabolism. Seminars in Cell & Developmental Biology, 23(6), 631–639. Puissegur, M. P., et al. (2011). miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death and Differentiation, 18(3), 465–478. Qiu, H. J., et al. (2016). MiR-769 promoted cell proliferation in human melanoma by suppressing GSK3B expression. Biomedicine & Pharmacotherapy, 82, 117–123. Qiu, Z., et al. (2015). MicroRNA-124 reduces the pentose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cells. Gastroenterology, 149(6), 1587-1598 e11. Qu, C., et al. (2020a). miR-128-3p contributes to mitochondrial dysfunction and induces apoptosis in glioma cells via targeting pyruvate dehydrogenase kinase 1. IUBMB Life, 72(3), 465–475. Qu, W., et al. (2016). miR-132 mediates a metabolic shift in prostate cancer cells by targeting Glut1. FEBS Open Bio, 6(7), 735–741. Qu, Z. Y., et al. (2020b). Potential suppressive functions of microRNA-504 in cervical cancer cells malignant process were achieved by targeting PAICS and regulating EMT. Archives of Gynecology and Obstetrics, 302(1), 173–182. Ranhotra, H. S. (2018). The estrogen-related receptors in metabolism and cancer: Newer insights. Journal of Receptor and Signal Transduction Research, 38(2), 95–100. Rayner, K. J., et al. (2010). MiR-33 contributes to the regulation of cholesterol homeostasis. Science, 328(5985), 1570–1573. Reinsborough, C. W., et al. (2021). BCDIN3D RNA methyltransferase stimulates Aldolase C expression and glycolysis through let-7 microRNA in breast cancer cells. Oncogene, 40(13), 2395–2406. Rohrig, F., & Schulze, A. (2016). The multifaceted roles of fatty acid synthesis in cancer. Nature Reviews Cancer, 16(11), 732–749. Romero-Cordoba, S. L., et al. (2018). Loss of function of miR-342-3p results in MCT1 over-expression and contributes to oncogenic metabolic reprogramming in triple negative breast cancer. Science and Reports, 8(1), 12252. Rottiers, V., & Naar, A. M. (2012). MicroRNAs in metabolism and metabolic disorders. Nature Reviews Molecular Cell Biology, 13(4), 239–250. Ru, P., et al. (2016). Feedback loop regulation of SCAP/SREBP-1 by miR-29 modulates EGFR signaling-driven glioblastoma growth. Cell Reports, 16(6), 1527–1535. Santasusagna, S., et al. (2018). miR-328 mediates a metabolic shift in colon cancer cells by targeting SLC2A1/GLUT1. Clinical and Translational Oncology, 20(9), 1161–1167. Schrem, H., Klempnauer, J., & Borlak, J. (2002). Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacological Reviews, 54(1), 129–158. Seeley, J. J., et al. (2018). Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature, 559(7712), 114–119. Selcuklu, S. D., et al. (2012). MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. Journal of Biological Chemistry, 287(35), 29516–29528. Sengupta, D., et al. (2020). Regulation of hepatic glutamine metabolism by miR-122. Molecular Metabolism, 34, 174–186. Sheng, W., et al. (2021). Curcumol inhibits the malignant progression of prostate cancer and regulates the PDK1/AKT/mTOR pathway by targeting miR9. Oncology Reports. https://doi.org/10.3892/or.2021.8197 Sheng, Y., et al. (2020). 3-Bromopyruvate inhibits the malignant phenotype of malignantly transformed macrophages and dendritic cells induced by glioma stem cells in the glioma microenvironment via miR-449a/MCT1. Biomedicine & Pharmacotherapy, 121, 109610. Shenoy, A., & Blelloch, R. H. (2014). Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nature Reviews Molecular Cell Biology, 15(9), 565–576. Shi, Y., et al. (2021). MTHFD2 promotes tumorigenesis and metastasis in lung adenocarcinoma by regulating AKT/GSK-3beta/beta-catenin signalling. Journal of Cellular and Molecular Medicine, 25(14), 7013–7027. Si, T., et al. (2021). microRNA-9-5p regulates the mitochondrial function of hepatocellular carcinoma cells through suppressing PDK4. Cancer Gene Therapy, 28(6), 706–718. Sikand, K., et al. (2012). Housekeeping gene selection advisory: Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-actin are targets of miR-644a. PLoS ONE, 7(10), e47510. Singh, A., et al. (2013). Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. The Journal of Clinical Investigation, 123(7), 2921–2934. Singh, R., et al. (2015). MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Science and Reports, 5, 17454. Sivanand, S., & Vander Heiden, M. G. (2020). Emerging roles for branched-chain amino acid metabolism in cancer. Cancer Cell, 37(2), 147–156. Snell, K. (1984). Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Advances in Enzyme Regulation, 22, 325–400. Song, L. R., et al. (2020). MicroRNA-195 functions as a tumor suppressor by directly targeting fatty acid synthase in malignant meningioma. World Neurosurg, 136, e355–e364. Stover, P., & Schirch, V. (1990). Serine hydroxymethyltransferase catalyzes the hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate. Journal of Biological Chemistry, 265(24), 14227–14233. Su, X., et al. (2020). miR-613 suppresses migration and invasion in esophageal squamous cell carcinoma via the targeting of G6PD. Experimental and Therapeutic Medicine, 19(4), 3081–3089. Sun, C., et al. (2018). MicroRNA-365 suppresses cell growth and invasion in esophageal squamous cell carcinoma by modulating phosphoserine aminotransferase 1. Cancer Management and Research, 10, 4581–4590. Sun, Q., et al. (2011). Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proceedings of the National Academy of Sciences of the United States of America, 108(10), 4129–4134. Sun, Q., et al. (2019b). LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Proliferation, 52(6), e12687. Sun, S., et al. (2021). Hsa_circ_0062682 promotes serine metabolism and tumor growth in colorectal cancer by regulating the miR-940/PHGDH axis. Frontiers in Cell and Developmental Biology, 9, 770006. Sun, X., & Zhang, L. (2017). MicroRNA-143 suppresses oral squamous cell carcinoma cell growth, invasion and glucose metabolism through targeting hexokinase 2. Bioscience Reports. https://doi.org/10.1042/BSR20160404 Sun, Y., et al. (2019a). The endonuclease APE1 processes miR-92b formation, thereby regulating expression of the tumor suppressor LDLR in cervical cancer cells. Therapeutic Advances in Medical Oncology, 11, 1758835919855859. Sun, Z., et al. (2015). MicroRNA-197 influences 5-fluorouracil resistance via thymidylate synthase in colorectal cancer. Clinical and Translational Oncology, 17(11), 876–883. Tanaka, H., et al. (2013). MicroRNA-183 upregulates HIF-1alpha by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. Journal of Neuro-Oncology, 111(3), 273–283. Tang, X., et al. (2021). Regulating COX10-AS1 / miR-142-5p / PAICS axis inhibits the proliferation of non-small cell lung cancer. Bioengineered, 12(1), 4643–4653. Tian, J., et al. (2016). MicroRNA-130b promotes lung cancer progression via PPARgamma/VEGF-A/BCL-2-mediated suppression of apoptosis. Journal of Experimental & Clinical Cancer Research, 35(1), 105. Tibbetts, A. S., & Appling, D. R. (2010). Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annual Review of Nutrition, 30, 57–81. Tiemin, P., et al. (2020). Dysregulation of the miR-148a-GLUT1 axis promotes the progression and chemoresistance of human intrahepatic cholangiocarcinoma. Oncogenesis, 9(2), 19. Tiwari, A., et al. (2014). MicroRNA-125a reduces proliferation and invasion of oral squamous cell carcinoma cells by targeting estrogen-related receptor alpha: Implications for cancer therapeutics. Journal of Biological Chemistry, 289(46), 32276–32290. Tomasetti, M., Neuzil, J., & Dong, L. (2014). MicroRNAs as regulators of mitochondrial function: Role in cancer suppression. Biochimica Et Biophysica Acta, 1840(4), 1441–1453. Tong, D., et al. (2020). MiR-22, regulated by MeCP2, suppresses gastric cancer cell proliferation by inducing a deficiency in endogenous S-adenosylmethionine. Oncogenesis, 9(11), 99. Tong, J. L., et al. (2011). MicroRNA 506 regulates expression of PPAR alpha in hydroxycamptothecin-resistant human colon cancer cells. FEBS Letters, 585(22), 3560–3568. Tsun, Z. Y., & Possemato, R. (2015). Amino acid management in cancer. Seminars in Cell & Developmental Biology, 43, 22–32. Valentino, A., et al. (2017). Deregulation of MicroRNAs mediated control of carnitine cycle in prostate cancer: Molecular basis and pathophysiological consequences. Oncogene, 36(43), 6030–6040. Vander Heiden, M. G., Cantley, L. C., & Thompson, C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science, 324(5930), 1029–1033. Vaupel, P., & Multhoff, G. (2021). Revisiting the Warburg effect: Historical dogma versus current understanding. Journal of Physiology, 599(6), 1745–1757. Wan, S., et al. (2021). MicroRNA-140-3p represses the proliferation, migration, invasion and angiogenesis of lung adenocarcinoma cells via targeting TYMS (thymidylate synthetase). Bioengineered, 12(2), 11959–11977. Wang, A., et al. (2021a). miR-206-G6PD axis regulates lipogenesis and cell growth in hepatocellular carcinoma cell. Anti-Cancer Drugs, 32(5), 508–516. Wang, B., et al. (2012). Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology, 56(1), 186–197. Wang, C. Q., et al. (2021b). Arbutin suppresses osteosarcoma progression via miR-338-3p/MTHFD1L and inactivation of the AKT/mTOR pathway. FEBS Open Bio, 11(1), 289–299. Wang, D., et al. (2019b). PYCR1 promotes the progression of non-small-cell lung cancer under the negative regulation of miR-488. Biomedicine & Pharmacotherapy, 111, 588–595. Wang, F., et al. (2018). MiR-155-5p inhibits PDK1 and promotes autophagy via the mTOR pathway in cervical cancer. International Journal of Biochemistry & Cell Biology, 99, 91–99. Wang, F., et al. (2020b). miR-6869-5p inhibits glioma cell proliferation and invasion via targeting PGK1. Mediators of Inflammation, 2020, 9752372. Wang, J., et al. (2019a). miR-9-5p inhibits pancreatic cancer cell proliferation, invasion and glutamine metabolism by targeting GOT1. Biochemical and Biophysical Research Communications, 509(1), 241–248. Wang, J., et al. (2020a). miR-202 functions as a tumor suppressor in hepatocellular carcinoma by targeting HK2. Oncology Letters, 19(3), 2265–2271. Wang, X., et al. (2021). Pyruvate dehydrogenase kinases (PDKs): an overview toward clinical applications. Bioscience Reports. https://doi.org/10.1042/BSR20204402 Wang, Y., et al. (2015). Transcriptional regulation of hepatic lipogenesis. Nature Reviews Molecular Cell Biology, 16(11), 678–689. Wang, Z., & Burke, P. A. (2013). The role of microRNAs in hepatocyte nuclear factor-4alpha expression and transactivation. Biochimica Et Biophysica Acta, 1829(5), 436–442. Wei, G., et al. (2020). USF1-mediated upregulation of lncRNA GAS6-AS2 facilitates osteosarcoma progression through miR-934/BCAT1 axis. Aging (albany NY), 12(7), 6172–6190. Weikum, E. R., Liu, X., & Ortlund, E. A. (2018). The nuclear receptor superfamily: A structural perspective. Protein Science, 27(11), 1876–1892. Wellen, K. E., & Thompson, C. B. (2012). A two-way street: Reciprocal regulation of metabolism and signalling. Nature Reviews Molecular Cell Biology, 13(4), 270–276. Wende, A. R., et al. (2005). PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: A mechanism for transcriptional control of muscle glucose metabolism. Molecular and Cellular Biology, 25(24), 10684–10694. Wu, K., et al. (2020b). microRNA-10b confers cisplatin resistance by activating AKT/mTOR/P70S6K signaling via targeting PPARgamma in esophageal cancer. Journal of Cellular Physiology, 235(2), 1247–1258. Wu, R., et al. (2020a). MiR-27a-3p targeting GSK3beta promotes triple-negative breast cancer proliferation and migration through Wnt/beta-catenin pathway. Cancer Management and Research, 12, 6241–6249. Wu, S., et al. (2016a). miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biology, 37(4), 5193–5202. Wu, X., et al. (2016b). miR-615-5p prevents proliferation and migration through negatively regulating serine hydromethyltransferase 2 (SHMT2) in hepatocellular carcinoma. Tumour Biology, 37(5), 6813–6821. Xiao, X., et al. (2016). The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Science and Reports, 6, 21735. Xie, H., et al. (2020). CASC9 plays a role in salivary adenoid cystic carcinoma in vitro by upregulation of ACLY. Oral Diseases. https://doi.org/10.1111/odi.13759 Xu, F., et al. (2020c). MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Science, 111(5), 1528–1541. Xu, G., et al. (2021a). Overexpression of miR-340 inhibits cell proliferation and induces apoptosis of human bladder cancer via targeting Glut-1. BMC Urology, 21(1), 168. Xu, H., et al. (2021b). MiR-1207-5p targets PYCR1 to inhibit the progression of prostate cancer. Biochemical and Biophysical Research Communications, 575, 56–64. Xu, Q., et al. (2015). Regulatory circuit of PKM2/NF-kappaB/miR-148a/152-modulated tumor angiogenesis and cancer progression. Oncogene, 34(43), 5482–5493. Xu, Q., et al. (2018b). Oviductus ranae protein hydrolysate (ORPH) inhibits the growth, metastasis and glycolysis of HCC by targeting miR-491-5p/PKM2 axis. Biomedicine & Pharmacotherapy, 107, 1692–1704. Xu, T., et al. (2019). MicroRNA-940 inhibits glioma progression by blocking mitochondrial folate metabolism through targeting of MTHFD2. American Journal of Cancer Research, 9(2), 250–269. Xu, W., et al. (2017). MicroRNA-330 inhibited cell proliferation and enhanced chemosensitivity to 5-fluorouracil in colorectal cancer by directly targeting thymidylate synthase. Oncology Letters, 13(5), 3387–3394. Xu, X., et al. (2020a). miR-215 inhibits colorectal cancer cell migration and invasion via targeting stearoyl-CoA desaturase. Computational and Mathematical Methods in Medicine, 2020, 5807836. Xu, Y., et al. (2018c). miR-27b-3p is involved in doxorubicin resistance of human anaplastic thyroid cancer cells via targeting peroxisome proliferator-activated receptor gamma. Basic & Clinical Pharmacology & Toxicology, 123(6), 670–677. Xu, Y., et al. (2020b). Circ_ASPH promotes cholangiocarcinoma growth and metastasis through the miR-581/ATP-binding cassette transporter G1 signaling pathway. Cancer Commun (lond), 40(10), 545–550. Xu, Y., et al. (2022). miRNA-199a-5p/SLC2A1 axis regulates glucose metabolism in non-small cell lung cancer. Journal of Cancer, 13(7), 2352–2361. Xu, Z., et al. (2018a). miR-122-5p inhibits the proliferation, invasion and growth of bile duct carcinoma cells by targeting ALDOA. Cellular Physiology and Biochemistry, 48(6), 2596–2606. Yamasaki, T., et al. (2013). Tumor-suppressive microRNA-1291 directly regulates glucose transporter 1 in renal cell carcinoma. Cancer Science, 104(11), 1411–1419. Yan, S., et al. (2015). MicroRNA-340 inhibits esophageal cancer cell growth and invasion by targeting phosphoserine aminotransferase 1. Cellular Physiology and Biochemistry, 37(1), 375–386. Yan, Y., et al. (2019). MicroRNA-33a-5p suppresses colorectal cancer cell growth by inhibiting MTHFD2. Clinical and Experimental Pharmacology and Physiology, 46(10), 928–936. Yang, H., et al. (2016b). MicroRNA-497 regulates cisplatin chemosensitivity of cervical cancer by targeting transketolase. American Journal of Cancer Research, 6(11), 2690–2699. Yang, H., et al. (2021). HK2 is a crucial downstream regulator of miR-148a for the maintenance of sphere-forming property and cisplatin resistance in cervical cancer cells. Frontiers in Oncology, 11, 794015. Yang, J., et al. (2016a). PFKL/miR-128 axis regulates glycolysis by inhibiting AKT phosphorylation and predicts poor survival in lung cancer. American Journal of Cancer Research, 6(2), 473–485. Yang, Q., et al. (2019). MiR-218-5p suppresses the killing effect of natural killer cell to lung adenocarcinoma by targeting SHMT1. Yonsei Medical Journal, 60(6), 500–508. Ye, F., et al. (2020b). LINC00963 confers oncogenic properties in glioma by regulating the miR-506/BCAT1 axis. Cancer Manag Res, 12, 2339–2351. Ye, J., et al. (2022). Long non-coding RNA TMPO-AS1 facilitates the progression of colorectal cancer cells via sponging miR-98-5p to upregulate BCAT1 expression. Journal of Gastroenterology and Hepatology, 37(1), 144–153. Ye, T., et al. (2020a). MicroRNA-16-1-3p represses breast tumor growth and metastasis by inhibiting PGK1-mediated Warburg effect. Frontiers in Cell and Developmental Biology, 8, 615154. Yi, W., et al. (2020). Bioengineered miR-328-3p modulates GLUT1-mediated glucose uptake and metabolism to exert synergistic antiproliferative effects with chemotherapeutics. Acta Pharmaceutica Sinica B, 10(1), 159–170. Yin, H., et al. (2021). HIF-1alpha downregulation of miR-433-3p in adipocyte-derived exosomes contributes to NPC progression via targeting SCD1. Cancer Science, 112(4), 1457–1470. Yoshino, H., et al. (2013). Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Science, 104(12), 1567–1574. Yu, G., et al. (2018). PKM2 functions as a potential oncogene and is a crucial target of miR-148a and miR-326 in thyroid tumorigenesis. American Journal of Translational Research, 10(6), 1793–1801. Yu, Q., et al. (2019). MicroRNA-214 suppresses cell proliferation and migration and cell metabolism by targeting PDK2 and PHF6 in hepatocellular carcinoma. Cell Biology International. https://doi.org/10.1002/cbin.11207 Yu, X., et al. (2020). ZHX2 inhibits SREBP1c-mediated de novo lipogenesis in hepatocellular carcinoma via miR-24-3p. The Journal of Pathology, 252(4), 358–370. Yu, Y., et al. (2019). LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncology, 15(33), 3831–3844. Yuan, B., et al. (2015). MiR-940 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer Science, 106(7), 819–824. Yuan, Y., et al. (2021). The miR-145-5p/CD36 pathway mediates PCB2-induced apoptosis in MCF-7 cells. Genes Genomics, 43(2), 161–171. Zeng, B., et al. (2019). The role of DNMT1/hsa-miR-124-3p/BCAT1 pathway in regulating growth and invasion of esophageal squamous cell carcinoma. BMC Cancer, 19(1), 609. Zeng, F., et al. (2022). Fatty acid beta-oxidation promotes breast cancer stemness and metastasis via the miRNA-328-3p-CPT1A pathway. Cancer Gene Therapy, 29(3–4), 383–395. Zhan, P., et al. (2021). miR-98-5p inhibits gastric cancer cell stemness and chemoresistance by targeting branched-chain aminotransferases 1. Life Sciences, 276, 119405. Zhang, D., et al. (2021a). MiR-489-3p reduced pancreatic cancer proliferation and metastasis by targeting PKM2 and LDHA involving glycolysis. Frontiers in Oncology, 11, 651535. Zhang, G., et al. (2021b). Energy stress-induced linc01564 activates the serine synthesis pathway and facilitates hepatocellular carcinogenesis. Oncogene, 40(16), 2936–2951. Zhang, H., et al. (2019a). miR-625-5p/PKM2 negatively regulates melanoma glycolysis state. Journal of Cellular Biochemistry, 120(3), 2964–2972. Zhang, H., et al. (2022). FASN targeted by miR-497–5p regulates cell behaviors in cervical cancer. Nutrition and Cancer. https://doi.org/10.1080/01635581.2022.2036351 Zhang, J., et al. (2014). Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Molecular Cell, 56(2), 205–218. Zhang, J., Han, C., & Wu, T. (2012a). MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology, 143(1), 246–56 e8. Zhang, K., et al. (2018). miR-9 regulates ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in melanoma. Molecular Carcinogenesis, 57(11), 1566–1576. Zhang, L., et al. (2015a). Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature, 527(7576), 100–104. Zhang, L. F., et al. (2015b). Suppression of miR-199a maturation by HuR is crucial for hypoxia-induced glycolytic switch in hepatocellular carcinoma. EMBO Journal, 34(21), 2671–2685. Zhang, N., et al. (2015c). MicroRNA-101 overexpression by IL-6 and TNF-alpha inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression. Experimental Cell Research, 336(1), 33–42. Zhang, N., et al. (2019b). SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2. Cell Death and Differentiation, 26(5), 843–859. Zhang, S., et al. (2017). PGC-1 alpha interacts with microRNA-217 to functionally regulate breast cancer cell proliferation. Biomedicine & Pharmacotherapy, 85, 541–548. Zhang, T., et al. (2022). MicroRNA-377-3p inhibits hepatocellular carcinoma growth and metastasis through negative regulation of CPT1C-mediated fatty acid oxidation. Cancer Metab, 10(1), 2. Zhang, Y., et al. (2012b). Downregulation of human farnesoid X receptor by miR-421 promotes proliferation and migration of hepatocellular carcinoma cells. Molecular Cancer Research, 10(4), 516–522. Zhao, X., Li, Y., & Zhou, Y. (2019b). MicroRNA-96-3p promotes metastasis of papillary thyroid cancer through targeting SDHB. Cancer Cell International, 19, 287. Zhao, Y., et al. (2019a). Targeted inhibition of MCT4 disrupts intracellular pH homeostasis and confers self-regulated apoptosis on hepatocellular carcinoma. Experimental Cell Research, 384(1), 111591. Zhao, Z., et al. (2020). miR-16-5p/PDK4-mediated metabolic reprogramming is involved in chemoresistance of cervical cancer. Molecular Therapy - Oncolytics, 17, 509–517. Zheng, J., et al. (2019). (125)I suppressed the Warburg effect viaregulating miR-338/PFKL axis in hepatocellular carcinoma. Biomedicine & Pharmacotherapy, 119, 109402. Zheng, K., et al. (2022). MicroRNA-206 inhibits HCV proliferation through depressing ACC1 lipid synthesis signalling pathway. Journal of Viral Hepatitis. https://doi.org/10.1111/jvh.13703 Zheng, S., et al. (2020). 1, 6-O, O-diacetylbritannilactone from inula britannica induces anti-tumor effect on oral squamous cell carcinoma via miR-1247-3p/LXRalpha/ABCA1 signaling. Oncotargets and Therapy, 13, 11097–11109. Zheng, W., et al. (2017). Inhibition of 6-phosphogluconate Dehydrogenase reverses cisplatin resistance in ovarian and lung cancer. Frontiers in Pharmacology, 8, 421. Zhong, D., et al. (2013). MicroRNA-1 and microRNA-206 suppress LXRalpha-induced lipogenesis in hepatocytes. Cellular Signalling, 25(6), 1429–1437. Zhong, X. Y., et al. (2012). MicroRNA-421 functions as an oncogenic miRNA in biliary tract cancer through down-regulating farnesoid X receptor expression. Gene, 493(1), 44–51. Zhou, P., Chen, W. G., & Li, X. W. (2015). MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. American Journal of Cancer Research, 5(6), 2056–2063. Zhou, X., et al. (2021). Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. Journal of Experimental & Clinical Cancer Research, 40(1), 124. Zhou, X., et al. (2021). miR-141–3p promotes the cisplatin sensitivity of osteosarcoma cell through targeting the Glutaminase (GLS)-mediated glutamine metabolism. Current Molecular Medicine. https://doi.org/10.2174/1566524021666211004112055 Zhu, W., Shao, Y., & Peng, Y. (2017). MicroRNA-218 inhibits tumor growth and increases chemosensitivity to CDDP treatment by targeting BCAT1 in prostate cancer. Molecular Carcinogenesis, 56(6), 1570–1577. Zhu, X., et al. (2020). A novel circular RNA hsa_circRNA_103809/miR-377-3p/GOT1 pathway regulates cisplatin-resistance in non-small cell lung cancer (NSCLC). BMC Cancer, 20(1), 1190. Zhuang, H., et al. (2019). Glycine decarboxylase induces autophagy and is downregulated by miRNA-30d-5p in hepatocellular carcinoma. Cell Death & Disease, 10(3), 192. Zu, X., et al. (2013). Chemical genetics of acetyl-CoA carboxylases. Molecules, 18(2), 1704–1719. This work was supported by grants from the National Natural Science Foundation of China (81930123, 81790252, 82121004). The National Natural Science Foundation of China (Grant no. 81930123); Innovative Research Group Project of the National Natural Science Foundation of China (Grant no. 81790252); the National Natural Science Foundation of China (Grant no. 82121004). Fudan University Shanghai Cancer Center and Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, 302 Rm., 7# Bldg., 270 Dong An Road, Shanghai, 200032, China Jie Ding, Yifan Wen, Xu Yuan & Xianghuo He Key Laboratory of Breast Cancer in Shanghai, Fudan University Shanghai Cancer Center, Fudan University, Shanghai, 200032, China Xianghuo He Shanghai Key Laboratory of Radiation Oncology, Fudan University Shanghai Cancer Center, Fudan University, Shanghai, 200032, China Xianghuo He Correspondence to Xianghuo He. Nothing to report. Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Ding, J., Wen, Y., Yuan, X. et al. MicroRNA-mediated reprogramming of glucose, fatty acid and amino acid metabolism in cancer. GENOME INSTAB. DIS. (2022). https://doi.org/10.1007/s42764-022-00078-x Received15 May 2022 Revised12 July 2022 Accepted05 August 2022 Published26 August 2022 DOIhttps://doi.org/10.1007/s42764-022-00078-x Anyone you share the following link with will be able to read this content: microRNAs Metabolic reprogramming Carbon sources CancerAbstract
Introduction
Roles of miRNAs in glucose metabolism
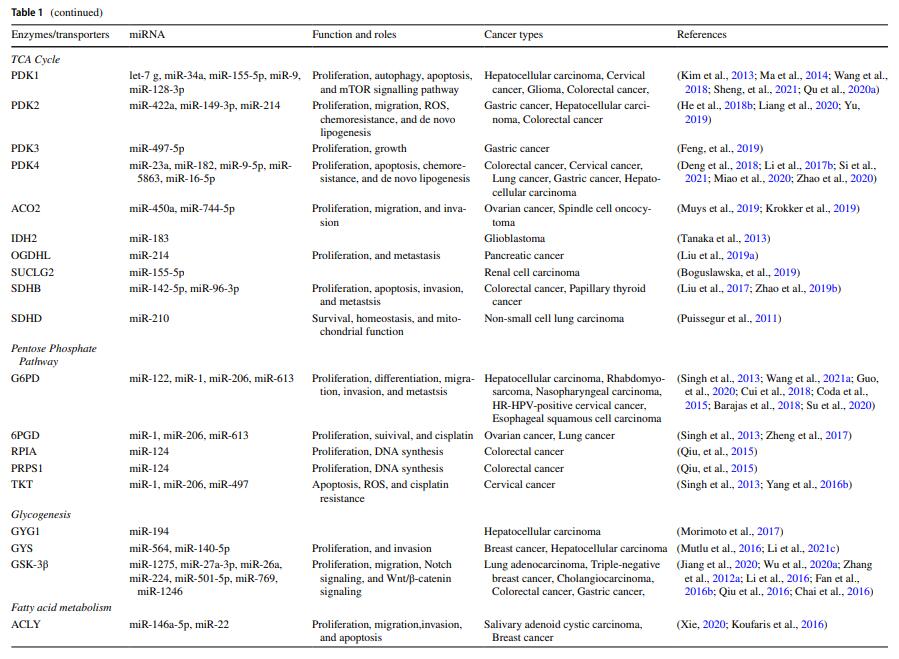
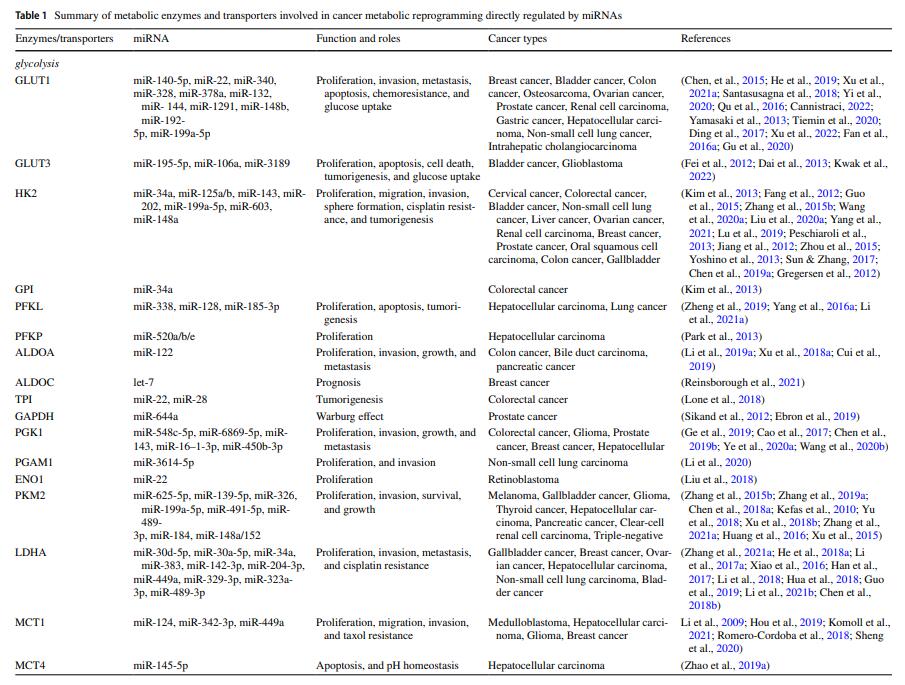
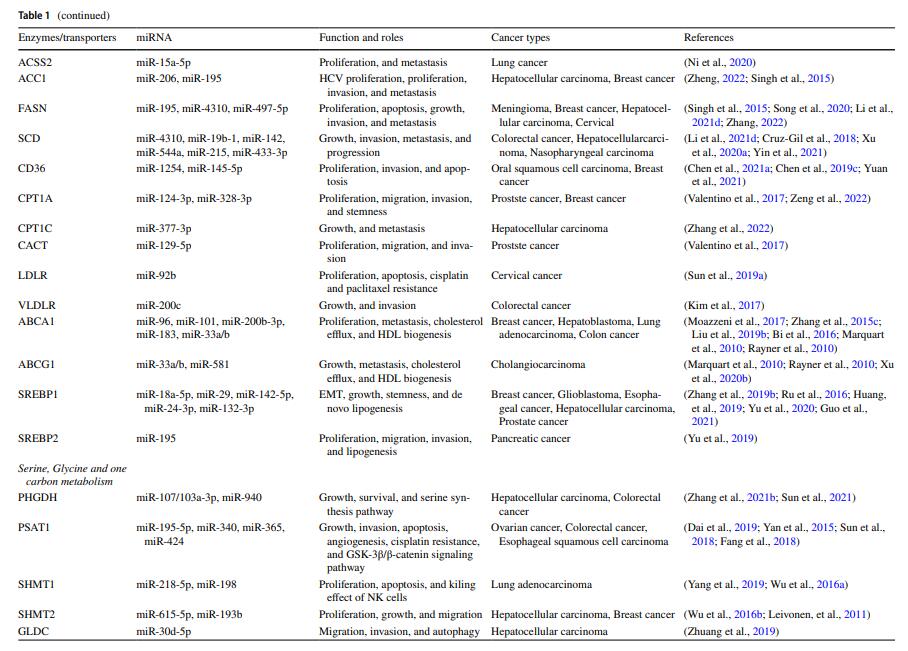
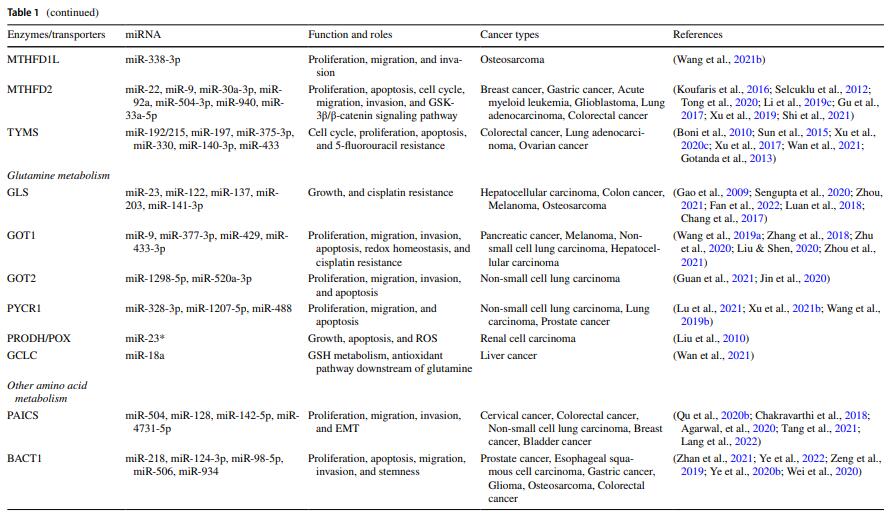

Roles of miRNAs in fatty acid metabolism
Roles of miRNAs in amino acid metabolism

Conclusive remarks
Availability of data and material
References
Acknowledgements
Funding
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Rights and permissions
About this article
Cite this article
Share this article
Keywords








用户登录
还没有账号?
立即注册