Drops in the cell ocean: new roles for non-coding RNAs in liquid–liquid phase separation
Review Article
Mingyue Li, Rick F. Thorne, Xu Dong Zhang, Mian Wu & Song Chen
Genome Instability & Disease (2022)
Abstract
Many types of membraneless organelles and related substructures occur in the nucleus and cytoplasm of cells, providing the essential framework for regulating innumerable biological activities. Fundamentally, these consist of RNA–protein (RNP) condensates formed by the process of liquid–liquid phase separation (LLPS). A salient attribute of these structures is their dynamic nature, a characteristic feature which dovetails with their essential roles in signal transduction and stress responses. In this regard, there is increasing evidence that non-coding RNAs serve as catalysts for the formation of LLPS structures. In this review, we summarize the current research in this field, focusing on how microRNAs, long non-coding RNAs, and circular RNAs contribute to the regulation of phase separation in different LLPS structures. In concert with this approach, we also shed new light onto the increasingly apparent role that phase separation plays in disease, particularly cancer. Finally, we lay out the challenges for this research and project how a deeper understanding of RNA-driven phase separation could help advance disease diagnosis and treatment.
Introduction
Membraneless organelles and organelle subdomains, alternatively termed ribonucleoprotein (RNP) granules in some literature, have largely been curiosities to many biologists. Examples of these structures in the cytoplasm include stress granules (SGs), processing bodies (P-bodies), and germline P-granules, while those in the nucleoplasm include the nucleolus, Cajal bodies, and paraspeckles (Banani et al., 2017; Shin & Brangwynne, 2017; Zhao & Zhang, 2020). While each organelle performs specialized cellular functions, they all have a common underlying structure and derivation, namely they consist of macromolecular condensates formed through liquid–liquid phase separation (LLPS). This process enables various biomolecular components to rapidly form condensates and, conversely, to dissolve rapidly (Banani et al., 2017; Shin & Brangwynne, 2017). At a fundamental level, membraneless organelles serve to enrich and concentrate reactants, with their isolated reactions protected from outside interferences, thus ensuring the order and efficiency of reactions (Guo et al., 2021). Notably, a major drawcard to a new generation of researchers being attracted to this field involves their role in signaling. Indeed, the dynamic condensation–dissolution of phase-separated droplets provides temporal control over the recruitment or release of signaling molecules, providing new insights into this central area of cell biology.
LLPS droplets primarily consist of proteins and nucleic acids driven together by weak multivalent interactions (Fig. 1). Such interactions involve intrinsically disordered regions (IDRs) or related low-complexity sequences (LCSs) in proteins with DNA or RNA molecules typically serving as scaffolds (Kato et al., 2012, Elbaum-Garfinkle et al., 2015; Nott et al., 2015, Murray, Kato et al. 2017, Alberti et al., 2019). The process of LLPS is typically sensitive to its surrounding environmental factors, such as temperature, pH, salt concentration, co-solute, osmotic pressure, and the concentration of other macromolecules (e.g., DNA, RNA, lipids, and proteins) (Yoo et al., 2019). Protein folding and post-translational modifications, such as phosphorylation (Larson et al., 2017), methylation (Qamar et al., 2018), ubiquitination (Dao, Kolaitis et al. 2018), and sumoylation (Qu, Wang et al., 2020a, 2020b), can also regulate LLPS along with nucleic acid modifications, such as m6A methylation in RNA (Ries et al., 2019). Notably, the component molecules within the droplets are mobile and readily undergo internal rearrangement and external exchange. A further characteristic of LLPS is that the droplets resume their spherical shape after touching and fusing with other droplets (Hyman et al., 2014), a feature which is important to verify experimentally when studying this phenomenon.
Fig. 1
Liquid–liquid phase separation. A schematic illustration of liquid–liquid phase separation (LLPS) involving lncRNAs’ solution composed of representative proteins with (yellow) and without intrinsic disordered regions (IDR) (blue) (left). A phase change state is triggered by lncRNA recruitment of IDR-containing proteins, forms a condensed liquid-like phase structure (right)
Full size imageAs the name suggests, RNA, including non-coding RNA (ncRNA), is implicitly involved in RNP condensation, cooperating with protein partners to drive LLPS formation and to modulate the properties of droplets (Huo et al., 2020; Luo et al., 2021). In this context, secondary structures in ncRNAs bind with RNA-binding proteins (RBPs) to regulate phase separation events (Fay & Anderson, 2018; Maharana et al., 2018; Pessina et al., 2019). Many RBPs involved in phase separation possess intrinsically disordered regions, low-complexity domains, and prion-like domains that are often enriched with polar and charged amino acids (Alberti et al., 2019). On the other hand, RNAs are highly negatively charged molecules and thus can promote RNP condensate formation through interacting with positively charged domains in RBPs (Henninger et al., 2021). Emerging evidence has also revealed that RNA function extends beyond molecular scaffolds, acting to decrease the viscosity of protein components and promote their diffusion (Elbaum-Garfinkle et al., 2015). Moreover, the primary function of several well-known ncRNAs involves LLPS. For instance, neurodegenerative-associated repetitive ncRNAs form phase-separated droplets in the nucleus (Jain & Vale, 2017), NEAT1 lncRNA plays a critical role as an architectural scaffold for paraspeckle proteins (Naganuma et al., 2012), and Xist lncRNA recruits chromatin inhibitory proteins to form X-chromosome inactivation (Cerase et al., 2019). All these examples are also notable because of their involvement in different disease states. However, understanding the full contribution of ncRNA-mediated phase separation to disease must be considered a work in progress with further investigations still needed.
Here, we review the current advances in the phase-separation field regarding the contributions of different ncRNAs (lncRNAs, microRNAs and circular RNAs) in RNP condensate formation and their involvement in normal and disease states, particularly cancer. We also discuss the potential of these ncRNAs as therapeutic targets to counteract phase separation-related pathologies. However, before this, we provide an entrée defining the different ncRNAs of interest and give a more comprehensive introduction to the different manifestations of LLPS that occur throughout the cell.
Non-coding RNAs
In general, non-coding RNAs are defined as being incapable of encoding proteins with a major division into housekeeping and regulatory classifications (Eddy, 2001). The housekeeping ncRNAs include ribosomal RNAs, transfer RNAs, and small nuclear and small nucleolar RNAs, while the main groupings of regulatory ncRNAs include small interfering RNA (siRNA), piwi-interacting RNA (piRNA), enhancer RNA (eRNA), promoter-associated RNA (PARs), Y RNA, microRNA (miRNA), circular RNA (circRNA, 200–10,000 nt), and long non-coding RNA (lncRNA, > 200 nt) (Fu, 2014; Mongelli et al., 2019, Zhang, Wu et al. 2019a, b). In this review, we primarily focus on the role of microRNAs and long non-coding RNAs, which have been most clearly documented for their roles in LLPS. CircRNAs also receive an honorable mention, although, as we discuss, the evidence for their widespread role in LLPS is currently pending.
Mature microRNAs of 21–23 nt length are generated from hairpin loop structures transcribed from discrete miRNA genes or from intronic sequences derived from pre-mRNAs. They are most famously known as the mediators of RNA interference, where their binding to target mRNAs, lncRNAs, and circRNAs is responsible for mediating gene silencing at post-transcriptional levels (Djuranovic et al., 2012; Dykes & Emanueli, 2017; Li et al., 2018). On the other hand, long non-coding RNAs comprise transcripts longer than 200 nt that are similarly transcribed to protein-coding genes by RNA polymerase type II machinery (Ma et al., 2013). Moreover, lncRNAs can bind with DNA, RNA, and protein to enact their diverse functional repertoire (Cai et al., 2020; Wu et al., 2016; Yin et al., 2012). Their roles encompass all levels of gene regulation including transcriptional (Beckedorff et al., 2013, Schertzer, Braceros et al. 2019, Ariel, Lucero et al., 2020), post-transcriptional (Kretz et al., 2013; Grelet et al., 2017, Guo, Ma et al., 2020), and post-translational regulation (Liu et al., 2020; Xing et al., 2018) along with other impressive contributions, including maintaining genome integrity (Dimitrova et al., 2014; Hu et al., 2018; Schmitt et al., 2016) and of relevance here, promoting the formation of cellular organelles (Leucci et al., 2016; Noh et al., 2016). CircRNAs are produced during the splicing of protein-coding or non-coding transcripts and typically arise from so-called back-splicing to produce covalently closed-loop structures (Noto et al., 2017; Salzman et al., 2012). Their size ranges from as little as 100 nt to greater than 10,000 nt. Much of the circRNA-associated literature shows that they serve as miRNA sponges (Sun et al., 2016) or competing endogenous RNAs (Huang et al., 2016), but emerging evidence shows that circRNAs are likely to rival lncRNAs in the diversity of their functions (Salzman, 2016).
Liquid–liquid phase separation
Liquid–liquid phase separation has recently emerged from the shadows to be regarded as a vital and ubiquitous process underlying the formation of membraneless droplets in the cell (Onoguchi-Mizutani & Akimitsu, 2022; Su et al., 2021). Increasing interest in this subject has promoted the application of more advanced technologies to study components of LLPS, which have enabled a better understanding of the different interaction states that exist between RNA and chromatin, RNA and protein, together with RNA–RNA and RNA–DNA hybrids (Guh et al., 2020). According to their localization, phase separation droplets have been classified into three spatial groups, namely plasma membrane clusters, cytoplasmic condensates, and nuclear bodies.
Examples of plasma membrane clusters include TCR clusters (Su et al., 2016), nephrin clusters (Banjade & Rosen, 2014), actin patches (Goode et al., 2015), focal adhesions (Shan et al., 2018), and synaptic densities (Zeng, Shang et al., 2016, Zeng, Chen et al., 2018), which respectively, are principally proposed to be involved in immune signal transduction, glomerular filtration barrier, endocytosis, cell migration, and neurotransmission. Cytoplasmic condensates include P-granules (Brangwynne et al., 2009), cGAS condensates (Du & Chen, 2018), Balbiani bodies (Boke et al., 2016), U bodies (Liu & Gall, 2007), P-bodies (Decker & Parker, 2012), RNA transport granules (Hofweber, Hutten et al., 2018), and stress granules (Yang, Mathieu et al., 2020), which are in variously involved in germ cell lineage maintenance, innate immune signaling, storage of snRNPs, mRNA decay, mRNA transport, and mRNA storage and translational regulation. Nuclear bodies variously consist of paraspeckles (Hennig et al., 2015), nuclear speckles (Spector & Lamond, 2011), cleavage bodies (Li et al., 2006), Cajal bodies (Handwerger et al., 2005; Nizami et al., 2010), PML bodies (Lallemand-Breitenbach & de The, 2010), PcG bodies (Pirrotta & Li, 2012), histone locus bodies (Duronio & Marzluff, 2017), OPT domains (Harrigan et al., 2011), and Gems (Cauchi, 2011). These sub-organelles are tasked with the storage of RNAs, mRNA splicing and processing, assembling spliceosomal small nuclear ribonucleoproteins, transcriptional regulation, transcriptional repression, and the processing of histone mRNAs.
Some readers might be surprised at the inclusion of many well-studied structures formed through LLPS. Nevertheless, this provides opportunities to reevaluate these structures from a new perspective. The collection of structures provided should also be considered far from complete, which serves to highlight a key point that the understanding of LLPS is still very limited. In addition, it is worth noting that the protein constituents of most of these LLPS structures are generally known, but the same cannot be said for the RNA molecules involved.
The following sections now look at what is currently known about the contribution of different types of ncRNAs in the regulation of biomolecular droplet formation.
MicroRNAs and LLPS
MiRNAs induce post-transcriptional gene silencing via RNAi by tethering the RNA-induced silencing complex (RISC) to partly complementary sequence motifs in target mRNAs predominantly found within their 3' untranslated regions (UTRs) (Bartel, 2009; Fabian & Sonenberg, 2012; Saj & Lai, 2011). This process is competitive and the presence of alternative RNA transcripts capable of binding the miRNAs, otherwise termed competing endogenous RNAs (ceRNAs), determines the degree of gene silencing. Consequently, aberrant changes in the expression of miRNAs or ceRNAs influence numerous cancer-relevant processes such as cell proliferation and differentiation, cell cycle progression, apoptosis, migration, and metabolism (Jansson & Lund, 2012). Providing a new perspective on the RNAi mechanism, it has been reported that key components of the RISC including Argonaute (Ago2), TNRC6B, GW182, Rck, and Mov10 undergo phase-separation into P-bodies (Eystathioy et al., 2003; Kulkarni et al., 2010; Liu et al., 2005; Sen & Blau, 2005; Sheu-Gruttadauria & MacRae, 2018). In the following section, we present specific examples showing the involvement of miRNAs in the formation and regulation of cellular RNP condensates and describe how this relates to the outcome of specific cellular processes.
Prior studies have revealed that miR-490-3p plays an important role in various cancer types through its regulation of different cellular mechanisms. For example, miR-490-3p inhibits the proliferation of A549 lung cancer cells by targeting CCND1 (Gu et al., 2014), and inhibits autophagy via targeting ATG7 in hepatocellular carcinoma (Ou et al., 2018), while serving as a tumor suppressor in colorectal cancer by inhibiting oncogenic VDAC1 expression (Liu et al., 2018). Moreover, miR-490-3p was also shown to suppress CDK1 expression in colon cancer; however, this inhibition is relieved after treating cells with the classical LLPS inhibitor 1, 6-hexanediol, indicating that miR-490-3p targeting of CDK1 occurred in an LLPS-dependent manner (Qin et al., 2021). Further investigations showed that miR-490-3p colocalized with Ago2 and TNRC6B, with fluorescence recovery after photobleaching experiments (FRAP), indicating that miR-490-3p functioned as part of the miRNA-induced silencing complex (miRISC) (Qin et al., 2021). Whether or not this functionality extends to the other reported miR-490-3p roles remains to be established. However, given the vast network of regulatory interactions affected by miRNAs, more examples seem inevitable.
Another broadly functional miRNA that has been found to be secreted by many cell types, including immune and cancer cells, is the exosome-specific miRNA, miR-223 (Jeffries et al., 2019). Much interest has been generated in secreted exosomes, in large part because of their long-range delivery of bioactive molecules such as miRNAs to other cells and tissues. The cargo borne by exosomes depends on the cell type and signaling state, but the underlying process has recently been suggested to occur in an LLPS manner, with sorting of miRNAs dependent upon RNA-binding proteins. The RBP YBX1 was shown to selectively concentrate miR-223 into in vitro condensates, while it formed a liquid-like condensate in cells that was required for the sorting of miR-223 rather than miR-190 or miR-144 into exosomes (Liu, Ma et al., 2021). The authors proposed that the YBX1 liquid-like condensates increase the local concentration of YBX1 molecules and bound RNA, thus directing the selective sorting miRNAs into exosomes and coupling RNP granules to RNA packing into exosomes. However, whether this mechanism reflects how other miRNAs are concentrated into exosomes, for example, miR-20b-5p in diabetic subjects (Katayama et al., 2019), remains to be seen.
LncRNAs and LLPS
Among the scientific literature, examples of the involvement of lncRNAs in LLPS are far more numerous than studies reported for other ncRNAs. LnRNAs have been primarily shown to serve as molecular scaffolds to engage RBPs to facilitate condensate formation (Guo et al., 2021; Luo et al., 2021, Onoguchi-Mizutani, Kirikae et al., 2021). This possibly reflects their longer transcript length of lncRNAs also facilitates the formation of secondary structures via their repetitive sequences which is an essential feature in their role in condensate formation. It can be noted from the following that many of the lncRNAs function in the nucleus where the net of the liquid-like structures has been termed the liquid nucleome (Strom & Brangwynne, 2019) with the lncRNAs functioning as scaffolds sometimes termed architectural RNAs (Chujo et al., 2016; Clemson et al., 2009). Apart from the nucleolus, most of the phase separation-induced structures are less conspicuous than those observed in the cytoplasm such as nuclear speckles and gems. Nevertheless, the wide array of nuclear condensate structures represents important compartments for many classical nuclear activities.
Sourced from an array of biological fields, the following collection of examples showcases our current understanding of the functional diversity of lncRNAs in their roles as mediators of phase separation. These include damage-induced long non-coding RNA (dilncRNA), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), glutamine insufficiency regulator of glutaminase lncRNA (GIRGL), human satellite III (HSATIII), NEAT1_2 lncRNA, NORAD lncRNA, xist lncRNA, PNCTR lncRNA, TNBL lncRNA, SNHG9 lncRNA, DIGIT lncRNA, and rIGSRNA (Table 1 and Fig. 2).
Table 1 LncRNAs and RBPs in regulating liquid–liquid phase separation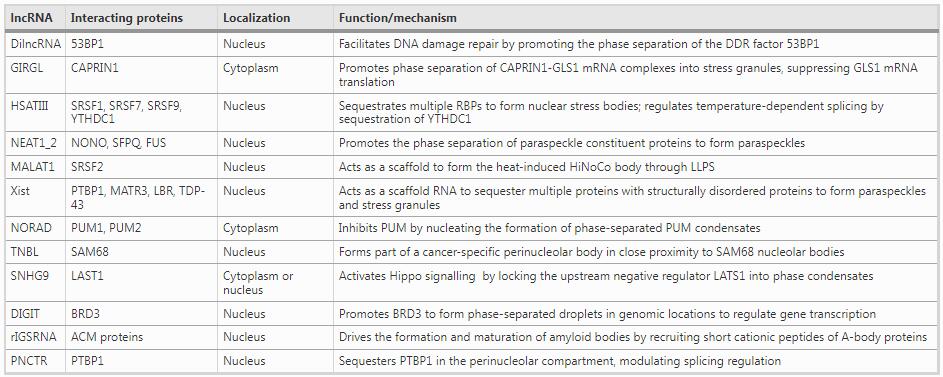
Full size table
Fig. 2
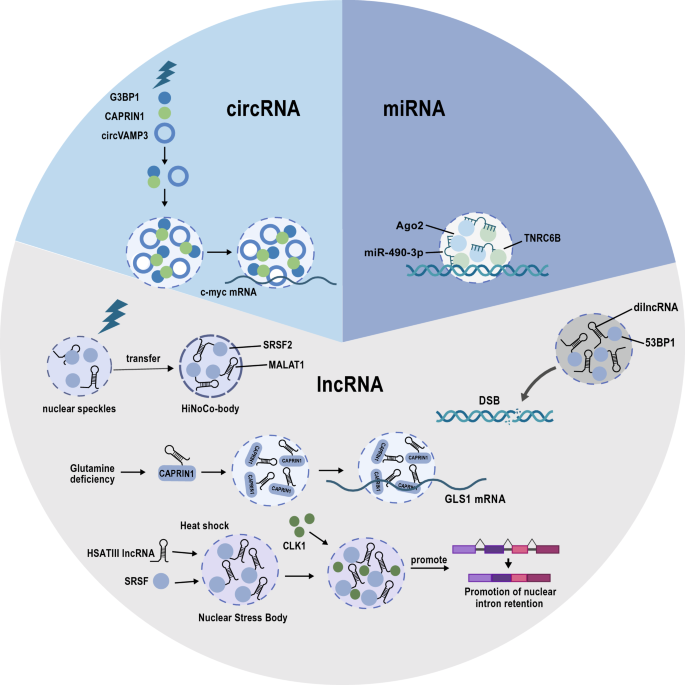
Non-coding RNAs are involved in liquid–liquid phase separation. MiR-490-3P binds to Ago2 and the scaffold protein TNRC6 to form the core of the miRNA-induced silencing complex (miRISC) in an LLPS-dependent manner (top right). CircVAMP3 can act as a molecular scaffold to recruit CAPRIN1-G3BP1 complex and drive their phase separation to inhibit the translation of c-Myc under stress conditions (top left). LncRNAs play a dominant role in the regulation of LLPS by recruiting RNA-binding proteins. Cellular stresses, including heat shock, glutamine deficiency, and DNA damage, lead to many lncRNA-LLPS structures such as stress granules, nuclear stress bodies, and HiNoCo bodies (bottom)
The damage-induced long non-coding RNA (dilncRNA) is induced in response to DNA-double strand breaks (DSB) to promote DNA damage repair (Michelini et al., 2017). Intriguingly, dilncRNA is transcribed by RNA polymerase II (RNAP II) directly at the DNA-damage-response (DDR) site. As revealed by in vitro experiments, DNA damage repair at the DSB is facilitated by dilncRNA in concert with a complete RNAP II pre-initiation complex (PIC) that promotes the phase separation of the DDR factor p53-binding protein 1 (53BP1) (Francia et al., 2016; Michelini et al., 2017; Pessina et al., 2019). It was also reported that unprocessed dilncRNA forms a DNA–RNA hybrid structure to recruit key proteins, such as BRCA1, BRCA2, and RAD51 to the DNA-double strand breaks site (D'Alessandro et al., 2018). Additionally, separate studies have indicated that the degradation of dilncRNA by the nuclear RNA exosome results in the accumulation of replication protein A (RPA) that binds to single-stranded DNA to initiate DNA repair by homologous recombination (Domingo-Prim et al., 2019).
The lncRNA GIRGL (glutamine insufficiency regulator of glutaminase lncRNA) was shown to contribute to cancer cell adaptation to fluctuating nutrient levels, a key feature of the tumor microenvironment (Wang, Cao et al., 2021a; b, c). When glutamine is limited, cells activate glutamine synthesis but unrestrained consumption of glutamine by glutaminase (mainly GLS1 in cancer cells) is detrimental to cell viability. Challenging cancer cells with glutamine deficiency resulted in the upregulation of GIRGL which promoted the homodimerization of CAPRIN1 to form a complex with GLS1 mRNA. This was demonstrated to involve the phase separation of CAPRIN1, indicating that GIRGL fundamentally functions by enhancing LLPS (Wang, Cao et al. 2021a; b, c). CAPRIN1, as mentioned above, is an RNA-binding protein that is commonly found in cytoplasmic stress granules (Kim, Payliss et al., 2021) which are known to impede the stability and translation of certain mRNAs (Decker & Parker, 2012). Indeed, GIRGL initiated CAPRIN1-mediated cytoplasmic stress granule formation to capture GLS1 mRNA with resulting suppression of GLS1 mRNA translation and consequent downregulation of GLS1. In turn, GIRGL promoted cell survival under highly limiting glutamine conditions.
Other lncRNAs associated with heat shock stress are the HSATIII lncRNAs transcribed by HSF1 (Heat Shock Transcription Factor 1) (Biamonti & Vourc’h, 2010), and interact with SR proteins (SRSF1, SRSF7, and SRSF9) accumulated in nuclear stress bodies, and these SR proteins could be immediately phosphorylated by CLK1 kinase in the recovery phase after heat shock (Ninomiya et al., 2020). Furthermore, during heat shock recovery, m6A-modified HSATIII lncRNAs repress the available nucleoplasmic levels of the m6A reader protein YTHDC1 via sequestration into nuclear stress bodies, in turn affecting m6A-dependent splicing eventsl (Ninomiya et al., 2021).
Arguably, the prototypical example of an lncRNA mediating liquid phase separation involves the nuclear-enriched abundant transcript 1 (NEAT1), which unlike many lncRNAs is highly conserved, at least in mammals (Guo et al., 2021). NEAT1 is alternatively termed ‘nuclear paraspeckle assembly transcript 1’ due to its connection to paraspeckles and other nuclear LLPS substructures found in non-chromatin regions (Fox et al., 2018; Hirose et al., 2019). NEAT1 is expressed as short and long isoforms, NEAT1_1 and NEAT1_2, respectively, that share a common 5’ sequence but with divergent 3’ ends. The latter in NEAT1_2 forms a triple helix and contains multiple protein interaction sites that support LLPS (Yamazaki et al., 2018). For example, NEAT1_2 interactions with the paraspeckle constituent proteins NONO (non-POU domain-containing octamer-binding protein) and SFPQ (splicing factor proline and glutamine-rich) serve to drive their phase separation to form paraspeckles (Imamura et al., 2014; Yamazaki et al., 2018). However, while NEAT1_1 is not found in paraspeckles, its expression does have regulatory significance for their formation, at least in the context of balancing stem cell pluripotency versus differentiation (Modic, Grosch et al. 2019). Critical to this functionality is the RBP TDP-43 which can repress the formation of paraspeckles in pluripotent stem cells through promoting polyadenylated Neat1_1. Conversely, TDP-43 can be sequestrated into paraspeckles by Neat1_2 (Wang et al., 2020a, 2020b), which is associated with stem cells exiting pluripotency (Modic, Grosch et al. 2019). Dysregulation of TDP-43–NEAT1 interactions has also been implicated in neurodegenerative pathologies such as familial amyotrophic lateral sclerosis (ALS). The assembly of nuclear bodies (NBs) involving LLPS involving NEAT1 and TDP-43 is proposed to be an adaptive mechanism in stressed neurons. However, the familial D169G mutation in TDP-43 affects the RNA recognition motif (RRM) bound by NEAT1, causing the failure of NB assembly, and redirecting TDP-43 to the cytoplasm to form stress granules (Wang et al., 2020a, 2020b).
A second NEAT lncRNA, nuclear-enriched abundant transcript 2 (NEAT2), is better known as MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) and is also one of the best characterized lncRNAs. Like NEAT1, MALAT1 is highly conserved and abundantly expressed in various cells and tissues (Arun, Aggarwal et al., 2020). A range of studies focusing on MALAT1 have revealed that it is not only relevant to cancer metastasis but contributes broad roles in a range of cell types, for instance, synaptogenesis in neurons (Bernard et al., 2010) and adipogenesis (Han et al., 2021). MALAT1 is retained in the nucleus where it primarily resides in nuclear speckles (Tripathi et al., 2010). These sub-organelles contain an array of proteins associated with RNA transcription and processing, including transcription and splicing factors, chromatin remodeling complexes, and various types of RNA molecules (Galganski et al., 2017). Notably, nuclear speckles coincide with certain actively transcribed chromatin regions, and these exhibit a clear overlap with genes regulated by MALAT1 (Engreitz et al., 2014). However, while most related studies acknowledge nuclear speckles as the site of action of MALAT1, most researchers have focused on the effects of MALAT1 on gene expression. Nonetheless, such activities are almost certainly related to the ability of MALAT1 to act as a scaffold to modify the composition of LLPS-related structures. For example, it has been shown that N6-methyladenosine-modified MALAT1 recruits the m6A reader, YTH-domain-containing protein 1 (YTHDC1) to nuclear speckles along with a broad complement of gene expression machinery. This process underlies the ability of MALAT1 to influence cancer cell motility through bringing nuclear speckles in proximity to key oncogenes, which enhances their expression (Wang, Liu et al., 2021). However, it must be remembered that while perturbations in MALAT1 function are exploited by cancer cells, MALAT1 also contributes to normal physiology. For instance, it was recently revealed that heat shock induces the translocation of MALAT1 from nuclear speckles [EE1] to a heat-inducible non-coding RNA containing nuclear body (HiNoCo body) formed through LLPS where it colocalizes with SRSF2 (a specific nuclear speckle marker protein) (Onoguchi-Mizutani, Kirikae et al., 2021). Notably, the knockout of MALAT1 reduces cell proliferation under heat shock stress, suggesting that HiNoCo body plays an important role in responding to heat shock (Onoguchi-Mizutani, Kirikae et al., 2021).
In mammals, the lncRNA X-inactive specific transcript (XIST) is considered the most essential gene for X-chromosome inactivation (XIC), but it also plays a critical role in cancer cell proliferation, differentiation, and genomic maintenance (). There are a number of lines of evidence to suggest that XIC represents a phase separation event (Cerase et al., 2019). Xist contains 6 conserved repeat sequences designated A–F, each with variable numbers and sizes of tandem repeats depending upon the species involved. These nucleotide repeats allow Xist to act as a scaffold RNA to sequester proteins, many of which are structurally disordered proteins that are components of paraspeckles and stress granules. For example, the human E repeat interacts with many RBPs, including PTBP1, MATR3, LBR, and TDP-43, proteins with a known propensity for phase separation (Cerase et al., 2019; Cirillo et al., 2016; Pandya-Jones et al., 2020). Importantly, there is experimental evidence that the binding partners of Xist can diffuse in a liquid-like manner, a key feature of LLPS (Cerase et al., 2019). Nonetheless, it will be crucial to better understand how Xist undergoes phase separation with its protein partners.
NORAD (Non-Coding RNA Activated by DNA Damage) has been associated with numerous processes related to carcinogenesis, including cell proliferation, apoptosis, invasion, and metastasis (Soghli et al., 2021). NORAD was originally reported to regulate genomic stability by sequestering PUMILIO (PUM) RNA-binding proteins. However, given that the cellular copy number of NORAD is greatly less than the total number of PUM-binding transcripts, it remained unclear how this lncRNA could effectively sequester the PUM proteins. However, a recent study provided the answer, demonstrating that NORAD can out-compete thousands of other transcripts via nucleating the formation of liquid-like cytoplasmic RNP condensates (Elguindy & Mendell, 2021). The authors found that NORAD-induced phase separation is driven by multivalent PUM-NORAD RNA-binding interactions and IDR-driven PUM–PUM interactions, supporting the regulation of PUM activity. It was further proposed that NORADs function in genomic stability in the RNP granules, in the same way that the interferon response pathway proteins IFIT1/2/3/5 coordinate regulation of DNA repair and replication (Lee et al., 2016; Luo et al., 2021; Tichon et al., 2016).
The tumor-associated NBL2 transcript or TNBL is an lncRNA transcribed from the NBL2 pericentromeric microsatellite region (Dumbovic et al., 2018). This region is noticeably hypomethylated in colon cancer cells with this status associated with TNBL biogenesis. Using advanced imaging techniques including STED, the authors showed that TNBL forms dense aggregates in interphase nuclei associated with NBL2 loci. Furthermore, these aggregates were found to cluster adjacent to SAM68 nuclear bodies, a perinucleolar substructure formed through LLPS (Mannen et al., 2016). Along with direct binding to SAM68, TNBL was shown to interact with other RBPs, namely NPM1 and CELF1. These proteins are known to be associated with alternative splicing, genome organization, and mRNA stability, respectively, with the authors suggesting that TNBL may function to disrupt nuclear organization in cancer cells. Notably, it was not formally demonstrated that the TNBL aggregates resulted from phase separation, and presently, this work has not been followed up in the literature.
The lncRNA SNHG9 (small nucleolar RNA host gene 9) has come to the attention of researchers, primarily because of its association with cancer and other disease states. However, there is no consensus as to whether SNHG9 dysregulation involves loss or overexpression as both occurrences have been reported. For instance, SNHG9 is downregulated in ovarian and non-small cell lung cancers (Chen et al., 2021; Wang et al., 2020a, 2020b), whereas its increased expression predicts worse outcomes in prostate cancer (Li et al., 2021a, 2021b). Moreover, most studies equate SNHG9 as acting through microRNA sponge mechanisms (Chen et al., 2021; Wang et al., 2020a, 2020b). However, a more intriguing role has been revealed involving LLPS where SNHG9 binds to lipids, specifically phosphatidic acid (PA) to prevent activation of the Salvador–Warts–Hippo (SWH or Hippo) pathway (Li et al., 2021a, 2021b). PA bridges the interaction between SNHG9 and the C-terminal domain of LATS1 (large tumor suppressor kinase 1), resulting in a phase separation event that restricts LATS1 from its normal function. Notably, LATS1 is the key upstream regulator that phosphorylates and destabilizes the Hippo pathway effector YAP1, and therefore, locking LATS1 into phase condensates effectively activates Hippo signaling. The significance of this process was elaborated in the context of breast cancer where silencing of SNHG9 inhibited tumor xenograft growth, and accordingly, in clinical tissues, there was a correlation between SNHG9, YAP activity, and cancer progression.
DIGIT is a conserved long non-coding RNA that plays an important role in specifying the endodermal differentiation route in embryonic stem cells (Daneshvar et al., 2016). The exact mechanism of how DIGIT contributes to endoderm differentiation was not initially known, but it was later shown that DIGIT interactions with the bromodomain-containing protein 3 (BRD3) were essential for directing endodermal differentiation. Notably, the addition of DIGIT in vitro or increasing cellular DIGIT expression promoted BRD3 to form phase-separated droplets (Daneshvar et al., 2020). Strikingly, these droplets were found to occupy genomic locations coincident with endodermal specification genes. DIGIT was shown to recruit BRD3 to acetylated histone H3 lysine 18 (H3K18ac) sites with their cooperation shown to be essential to regulate transcription factor occupancy at these sites. The authors reflected that protein-lncRNA phase-separated condensates may broadly act as regulators of different transcriptional programs other than endodermal differentiation.
Ribosomal intergenic spacer RNAs (rIGSRNAs) consist of repeated clusters cytosine/uracil or adenosine/guanine (AG) transcribed from the ~ 28 kbp ribosomal DNA intergenic spacer region (Audas et al., 2012). The rIGSRNAs are induced by cellular stress and play an intriguing role in the maturation of amyloid bodies (Knowles et al., 2014). Otherwise known as A-bodies, these are membraneless nuclear structures consisting of protein aggregates in a fibrillar amyloid-like state (Audas et al., 2016; Woodruff et al., 2018). The low-complexity repeat sequences in rIGSRNAs interact with cationic peptide sequences in the nucleolus, creating multiple LLPS-like droplets that act as seeds for A-body formation. In response to acidosis or heat shock stress, the reported role of A-bodies involves triggering cells to enter a dormant state (Audas et al., 2016; Wang et al., 2018). Notably, in contrast to classical phase separation structures, mature A-bodies are not dynamic, although it has been reported that they can be reversed by heat shock chaperones (Audas et al., 2016), suggesting that they represent a physiological adaptative response. Further distinguishing A-bodies from pathogenic amyloids, A-bodies are formed by a heterogeneous collective of more than 100 proteins (Audas et al., 2016) compared to a dominant protein constituent such as the Huntingtin protein in Huntington's disease.
Another lncRNA derived from the ribosomal DNA intergenic spacer region is the pyrimidine-rich non-coding transcript or PNCTR. This large ~ 10-kb transcript contains literally thousands of binding motifs for the RBP polypyrimidine tract-binding protein 1 (PTBP1). The latter protein is well known to be involved in regulating pre-mRNA processing and RNA splicing (Keppetipola et al., 2012) with the function of PTBP1 implicated in the tumorigenicity of many cancers (Yap et al., 2018). Notably, PNCTR is differentially overexpressed in cancer versus normal cells and is generally more abundant than rIGSRNAs, at least under tonic conditions (Yap et al., 2018). Because of the increased PNCTR expression in cancer cells, PTBP1 is sequestered to the perinucleolar compartment (PNC) in an LLPS manner. The resulting modulation of the cellular localization of PTBP1 and effects on splicing appear to have important functional consequences, since suppressing PNCTR expression induces apoptosis in cancer cells. Together, these studies show that multivalent binding between lncRNAs and RBPs is an effective mechanism of regulating disease-specific alternative splicing (Luo et al., 2021; Yap et al., 2018).
CircRNAs and LLPS
Over the past decade, circular RNAs (circRNAs) have emerged as a novel class of non-coding RNA molecules. These covalently linked ring structures are generated by back-splicing events in pre-RNA transcripts and are further distinguished from their linear counterparts through lacking traditional regulatory elements such 5′ and 3′ UTRs and polyadenylated tails (Suzuki & Tsukahara, 2014). CircRNAs can be generated from both coding and non-coding genes, and can consist of both exonic and intronic sequences, although exon-only transcripts largely prevail. They have been shown to be important players in numerous cellular functions that contribute to various physiological and pathological processes. The underlying mechanisms responsible are diverse with circRNAs reported to engage in miRNA sponging along with various protein binding activities, ranging from interactions with RNA-binding proteins through to effects on regulating transcription, splicing, and protein translation (Kristensen et al., 2022; Su et al., 2019). Breaking the definition of non-coding RNAs, they can even serve as templates for translation into peptides (Liu & Chen, 2022). Hence, given some of the previous examples discussed in this review together with the explosion of circRNA research, it would be surprising if circRNAs did not engage in phase separation. Nevertheless, published studies in this area are currently rare.
One prominent example involving a circRNA which can drive phase separation is circVAMP3 (Chen et al., 2022). CircVAMP3 is derived from the back-splicing of exons 3 and 4 in VAMP3, and like the vast majority of mature circRNAs, it is mainly located in the cytoplasm. Here, it engages with the complex of familiar LLPS actors, namely CAPRIN1 and G3BP1 through direct interactions with CAPRIN1. Notably, functional analyses showed that circVAMP3 inhibited the proliferation and metastasis of hepatocellular carcinoma (HCC) cells both in vitro and in vivo. Further investigations revealed the molecular mechanism underlying its carcinostatic role whereby circVAMP3 promotes the formation of cytoplasmic condensates of CAPRIN1 and G3BP1, which act to capture c-Myc mRNA. This process serves to inhibit the translation of c-Myc under stress conditions, which explains how circVAMP3 inhibits cell proliferation in HCC (Chen et al., 2022).
Given that circRNAs make up a major component of the cellular ncRNA complement, it appears likely that their role in LLPS events is underrepresented in the literature. Hence, more studies of this nature are eagerly anticipated.
Concluding remarks
This review summarizes the contributions of specific non-coding RNAs to LLPS and their consequent regulation of different phase-separation-related structures within cells. The ncRNAs do not act alone in these processes but engage with a plethora of different molecules, namely proteins, other RNAs, lipids, and chromatin. And, while the current list of ncRNAs involved in LLPS is presently limited, this does not necessarily mean their involvement is rare. Rather, many of the functions currently ascribed to ncRNAs likely involve elements of LLPS but such details remain hidden from the usual laboratory workflows. However, as LLPS research expands, it is expected that substantially more ncRNAs will be uncovered as key regulators of phase separation events. Moreover, researcher awareness of LLPS is also important, which is being helped by the publication of enticing new studies in high-profile journals.
Importantly, there are benefits for better understanding LLPS across all biomedical disciplines, particularly given the emerging evidence linking aberrant phase separation with a spectrum of human diseases. For example, certain neurodegenerative diseases are manifestations of phase separation gone awry. As already mentioned, aberrant mislocalization and aggregation of TDP-43 underlies familial ALS (Arnold et al., 2013; Suk & Rousseaux, 2020), but similar processes occur with other RBPs, such as FUS, which is involved in frontotemporal dementia (FTD) (Lopez-Erauskin, Tadokoro et al., 2018).
It is also pertinent to consider what can be done with this new knowledge. Are ncRNAs likely to be better treatment targets than proteins for example? New technologies to manipulate RNAs are appearing in clinical trials with some treatments being approved (Zogg, Singh et al. 2022). The answer will therefore lie in the context of the mechanism or pathway involved, but it now seems possible to add RNA therapeutics into the clinical pallet. In any case, the technologies to measure different RNA species are already embedded in many clinical laboratories, providing a clear path to implement ncRNAs as diagnostic biomarkers.
Finally, there are some important challenges to consider on the journey to advance our knowledge of ncRNAs and LLPS events in the cell. Determining why particular ncRNAs are associated with different phase separation droplets requires more than defining their cellular location. Rather, careful work is required to establish which factors are involved in phase droplet assembly and disassembly. Adding complexity to this task is the dynamic nature of LLPS and the impact of environmental stressors, which, as shown by many examples in this review, is a fundamental regulator of LLPS. Other challenges to be considered are the vast number of different ncRNA transcripts, both at the individual gene level and as alternatively spliced transcripts. As revealed by the example of NEAT1, splice variants could conceivably represent transcripts either with or without LLPS-initiating properties. Also, post-transcriptional modifications of ncRNAs, such as the m6A modification in MALAT1, are likely to be as important as post-translational modifications in proteins in regulating their activities. There is also a conceptual issue with the terms used for the different phase-separated entities in cells, especially those within the nucleus. As this field gathers momentum, it may be time to workshop naming conventions to help clarify this field to expert and novice researchers alike.
Data avaiability statement
No research data is included in this report. Please consult the authors of the individual studies for access to research data and materials.
References
Alberti, S., Gladfelter, A., & Mittag, T. (2019). Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell, 176(3), 419–434.
Ariel, F., Lucero, L., Christ, A., Mammarella, M. F., Jegu, T., Veluchamy, A., Mariappan, K., Latrasse, D., Blein, T., Liu, C., Benhamed, M., & Crespi, M. (2020). R-loop mediated trans action of the APOLO long noncoding RNA. Molecular Cell, 77(5), 1055–1065. e1054.
Arnold, E. S., Ling, S. C., Huelga, S. C., Lagier-Tourenne, C., Polymenidou, M., Ditsworth, D., Kordasiewicz, H. B., McAlonis-Downes, M., Platoshyn, O., Parone, P. A., Da Cruz, S., Clutario, K. M., Swing, D., Tessarollo, L., Marsala, M., Shaw, C. E., Yeo, G. W., & Cleveland, D. W. (2013). ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proceedings of the National Academy of Sciences USA, 110(8), E736-745.
Arun, G., Aggarwal, D., & Spector, D. L. (2020). MALAT1 long non-coding RNA: functional implications. Noncoding RNA, 6(2), 22.
Audas, T. E., Audas, D. E., Jacob, M. D., Ho, J. J., Khacho, M., Wang, M., Perera, J. K., Gardiner, C., Bennett, C. A., Head, T., Kryvenko, O. N., Jorda, M., Daunert, S., Malhotra, A., Trinkle-Mulcahy, L., Gonzalgo, M. L., & Lee, S. (2016). Adaptation to stressors by systemic protein amyloidogenesis. Developmental Cell, 39(2), 155–168.
Audas, T. E., Jacob, M. D., & Lee, S. (2012). Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Molecular Cell, 45(2), 147–157.
Banani, S. F., Lee, H. O., Hyman, A. A., & Rosen, M. K. (2017). Biomolecular condensates: Organizers of cellular biochemistry. Nature Reviews Molecular Cell Biology, 18(5), 285–298.
Banjade, S., & Rosen, M. K. (2014). Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife, 3, e04123.
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136(2), 215–233.
Beckedorff, F. C., Ayupe, A. C., Crocci-Souza, R., Amaral, M. S., Nakaya, H. I., Soltys, D. T., Menck, C. F., Reis, E. M., & Verjovski-Almeida, S. (2013). The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genetics, 9(8), e1003705.
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., Zhang, M. Q., Sedel, F., Jourdren, L., Coulpier, F., Triller, A., Spector, D. L., & Bessis, A. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO Journal, 29(18), 3082–3093.
Biamonti, G., & Vourc’h, C. (2010). Nuclear stress bodies. Cold Spring Harbor Perspectives in Biology, 2(6), a000695.
Boke, E., Ruer, M., Wuhr, M., Coughlin, M., Lemaitre, R., Gygi, S. P., Alberti, S., Drechsel, D., Hyman, A. A., & Mitchison, T. J. (2016). Amyloid-like self-assembly of a cellular compartment. Cell, 166(3), 637–650.
Brangwynne, C. P., Eckmann, C. R., Courson, D. S., Rybarska, A., Hoege, C., Gharakhani, J., Julicher, F., & Hyman, A. A. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science, 324(5935), 1729–1732.
Cai, Z., Cao, C., Ji, L., Ye, R., Wang, D., Xia, C., Wang, S., Du, Z., Hu, N., Yu, X., Chen, J., Wang, L., Yang, X., He, S., & Xue, Y. (2020). RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature, 582(7812), 432–437.
Cauchi, R. J. (2011). Gem formation upon constitutive Gemin3 overexpression in Drosophila. Cell Biology International, 35(12), 1233–1238.
Cerase, A., Armaos, A., Neumayer, C., Avner, P., Guttman, M., & Tartaglia, G. G. (2019). Phase separation drives X-chromosome inactivation: A hypothesis. Nature Structural & Molecular Biology, 26(5), 331–334.
Chen, G. Y., Zhang, Z. S., Chen, Y., & Li, Y. (2021). Long non-coding RNA SNHG9 inhibits ovarian cancer progression by sponging microRNA-214-5p. Oncology Letters, 21(2), 80.
Chen, S., Cao, X., Zhang, J., Wu, W., Zhang, B., & Zhao, F. (2022). circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-Myc translation. Advanced Science (weinh), 9(8), e2103817.
Chujo, T., Yamazaki, T., & Hirose, T. (2016). Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochimica Et Biophysica Acta, 1859(1), 139–146.
Cirillo, D., Blanco, M., Armaos, A., Buness, A., Avner, P., Guttman, M., Cerase, A., & Tartaglia, G. G. (2016). Quantitative predictions of protein interactions with long noncoding RNAs. Nature Methods, 14(1), 5–6.
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., & Lawrence, J. B. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Molecular Cell, 33(6), 717–726.
D’Alessandro, G., Whelan, D. R., Howard, S. M., Vitelli, V., Renaudin, X., Adamowicz, M., Iannelli, F., Jones-Weinert, C. W., Lee, M., Matti, V., Lee, W. T. C., Morten, M. J., Venkitaraman, A. R., Cejka, P., Rothenberg, E., & d’Adda di Fagagna, F. (2018). BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nature Communications, 9(1), 5376.
Daneshvar, K., Ardehali, M. B., Klein, I. A., Hsieh, F. K., Kratkiewicz, A. J., Mahpour, A., Cancelliere, S. O. L., Zhou, C., Cook, B. M., Li, W., Pondick, J. V., Gupta, S. K., Moran, S. P., Young, R. A., Kingston, R. E., & Mullen, A. C. (2020). lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nature Cell Biology, 22(10), 1211–1222.
Daneshvar, K., Pondick, J. V., Kim, B. M., Zhou, C., York, S. R., Macklin, J. A., Abualteen, A., Tan, B., Sigova, A. A., Marcho, C., Tremblay, K. D., Mager, J., Choi, M. Y., & Mullen, A. C. (2016). DIGIT Is a conserved long noncoding RNA that regulates GSC expression to control definitive endoderm differentiation of embryonic stem cells. Cell Reports, 17(2), 353–365.
Dao, T. P., Kolaitis, R. M., Kim, H. J., O’Donovan, K., Martyniak, B., Colicino, E., Hehnly, H., Taylor, J. P., & Castaneda, C. A. (2018). Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Molecular Cell, 69(6), 965–978. e966.
Decker, C. J., & Parker, R. (2012). P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harbor Perspectives in Biology, 4(9), a012286.
Dimitrova, N., Zamudio, J. R., Jong, R. M., Soukup, D., Resnick, R., Sarma, K., Ward, A. J., Raj, A., Lee, J. T., Sharp, P. A., & Jacks, T. (2014). LincRNA-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Molecular Cell, 54(5), 777–790.
Djuranovic, S., Nahvi, A., & Green, R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science, 336(6078), 237–240.
Domingo-Prim, J., Endara-Coll, M., Bonath, F., Jimeno, S., Prados-Carvajal, R., Friedländer, M. R., Huertas, P., & Visa, N. (2019). EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks. Nature Communications, 10(1), 2135.
Du, M., & Chen, Z. J. (2018). DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science, 361(6403), 704–709.
Dumbovic, G., Biayna, J., Banús, J., Samuelsson, J., Roth, A., Diederichs, S., Alonso, S., Buschbeck, M., Perucho, M., & Forcales, S. V. (2018). A novel long non-coding RNA from NBL2 pericentromeric macrosatellite forms a perinucleolar aggregate structure in colon cancer. Nucleic Acids Research, 46(11), 5504–5524.
Duronio, R. J., & Marzluff, W. F. (2017). Coordinating cell cycle-regulated histone gene expression through assembly and function of the histone locus body. RNA Biology, 14(6), 726–738.
Dykes, I. M., & Emanueli, C. (2017). Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics, Proteomics & Bioinformatics, 15(3), 177–186.
Eddy, S. R. (2001). Non-coding RNA genes and the modern RNA world. Nature Reviews Genetics, 2(12), 919–929.
Elbaum-Garfinkle, S., Kim, Y., Szczepaniak, K., Chen, C.C.-H., Eckmann, C. R., Myong, S., & Brangwynne, C. P. (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proceedings of the National Academy of Sciences of the United States of America, 112(23), 7189–7194.
Elguindy, M. M., & Mendell, J. T. (2021). NORAD-induced pumilio phase separation is required for genome stability. Nature, 595(7866), 303–308.
Engreitz, J. M., Sirokman, K., McDonel, P., Shishkin, A. A., Surka, C., Russell, P., Grossman, S. R., Chow, A. Y., Guttman, M., & Lander, E. S. (2014). RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell, 159(1), 188–199.
Eystathioy, T., Jakymiw, A., Chan, E. K., Seraphin, B., Cougot, N., & Fritzler, M. J. (2003). The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA, 9(10), 1171–1173.
Fabian, M. R., & Sonenberg, N. (2012). The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nature Structural & Molecular Biology, 19(6), 586–593.
Fay, M. M., & Anderson, P. J. (2018). The Role of RNA in biological phase separations. Journal of Molecular Biology, 430(23), 4685–4701.
Fox, A. H., Nakagawa, S., Hirose, T., & Bond, C. S. (2018). Paraspeckles: where long noncoding RNA meets phase separation. Trends in Biochemical Sciences, 43(2), 124–135.
Francia, S., Cabrini, M., Matti, V., Oldani, A., & d’Adda di Fagagna, F. (2016). DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. Journal of Cell Science, 129(7), 1468–1476.
Fu, X. D. (2014). Non-coding RNA: A new frontier in regulatory biology. National Science Review, 1(2), 190–204.
Galganski, L., Urbanek, M. O., & Krzyzosiak, W. J. (2017). Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Research, 45(18), 10350–10368.
Goode, B. L., Eskin, J. A., & Wendland, B. (2015). Actin and endocytosis in budding yeast. Genetics, 199(2), 315–358.
Grelet, S., Link, L. A., Howley, B., Obellianne, C., Palanisamy, V., Gangaraju, V. K., Diehl, J. A., & Howe, P. H. (2017). A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nature Cell Biology, 19(9), 1105–1115.
Gu, H., Yang, T., Fu, S., Chen, X., Guo, L., & Ni, Y. (2014). MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells by targeting CCND1. Biochemical and Biophysical Research Communications, 444(1), 104–108.
Guh, C. Y., Hsieh, Y. H., & Chu, H. P. (2020). Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. Journal of Biomedical Science, 27(1), 44.
Guo, C. J., Ma, X. K., Xing, Y. H., Zheng, C. C., Xu, Y. F., Shan, L., Zhang, J., Wang, S., Wang, Y., Carmichael, G. G., Yang, L., & Chen, L. L. (2020). Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell, 181(3), 621–636. e622.
Guo, Q., Shi, X., & Wang, X. (2021). RNA and liquid-liquid phase separation. Noncoding RNA Research, 6(2), 92–99.
Han, J., Shen, L., Zhan, Z., Liu, Y., Zhang, C., Guo, R., Luo, Y., Xie, Z., Feng, Y., & Wu, G. (2021). The long noncoding RNA MALAT1 modulates adipose loss in cancer-associated cachexia by suppressing adipogenesis through PPAR-gamma. Nutrition & Metabolism (london), 18(1), 27.
Handwerger, K. E., Cordero, J. A., & Gall, J. G. (2005). Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Molecular Biology of the Cell, 16(1), 202–211.
Harrigan, J. A., Belotserkovskaya, R., Coates, J., Dimitrova, D. S., Polo, S. E., Bradshaw, C. R., Fraser, P., & Jackson, S. P. (2011). Replication stress induces 53BP1-containing OPT domains in G1 cells. Journal of Cell Biology, 193(1), 97–108.
Hennig, S., Kong, G., Mannen, T., Sadowska, A., Kobelke, S., Blythe, A., Knott, G. J., Iyer, K. S., Ho, D., Newcombe, E. A., Hosoki, K., Goshima, N., Kawaguchi, T., Hatters, D., Trinkle-Mulcahy, L., Hirose, T., Bond, C. S., & Fox, A. H. (2015). Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. Journal of Cell Biology, 210(4), 529–539.
Henninger, J. E., Oksuz, O., Shrinivas, K., Sagi, I., LeRoy, G., Zheng, M. M., Andrews, J. O., Zamudio, A. V., Lazaris, C., Hannett, N. M., Lee, T. I., Sharp, P. A., Cissé, II, Chakraborty A. K., Young R. A. (2021). RNA-mediated feedback control of transcriptional condensates. Cell, 184(1): 207-225. e224
Hirose, T., Yamazaki, T., & Nakagawa, S. (2019). Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdisciplinary Reviews RNA, 10(6), e1545.
Hofweber, M., Hutten, S., Bourgeois, B., Spreitzer, E., Niedner-Boblenz, A., Schifferer, M., Ruepp, M. D., Simons, M., Niessing, D., Madl, T., & Dormann, D. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell, 173(3), 706–719. e713.
Hu, W. L., Jin, L., Xu, A., Wang, Y. F., Thorne, R. F., Zhang, X. D., & Wu, M. (2018). GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nature Cell Biology, 20(4), 492–502.
Huang, M., Zhong, Z., Lv, M., Shu, J., Tian, Q., & Chen, J. (2016). Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget, 7(30), 47186–47200.
Huo, X., Ji, L., Zhang, Y., Lv, P., Cao, X., Wang, Q., Yan, Z., Dong, S., Du, D., Zhang, F., Wei, G., Liu, Y., & Wen, B. (2020). The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Molecular Cell, 77(2), 368-383.e367.
Hyman, A. A., Weber, C. A., & Julicher, F. (2014). Liquid-liquid phase separation in biology. Annual Review of Cell and Developmental Biology, 30, 39–58.
Imamura, K., Imamachi, N., Akizuki, G., Kumakura, M., Kawaguchi, A., Nagata, K., Kato, A., Kawaguchi, Y., Sato, H., Yoneda, M., Kai, C., Yada, T., Suzuki, Y., Yamada, T., Ozawa, T., Kaneki, K., Inoue, T., Kobayashi, M., Kodama, T., … Akimitsu, N. (2014). Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Molecular Cell, 53(3), 393–406.
Jain, A., & Vale, R. D. (2017). RNA phase transitions in repeat expansion disorders. Nature, 546(7657), 243–247.
Jansson, M. D., & Lund, A. H. (2012). MicroRNA and cancer. Molecular Oncology, 6(6), 590–610.
Jeffries, J., Zhou, W., Hsu, A. Y., & Deng, Q. (2019). miRNA-223 at the crossroads of inflammation and cancer. Cancer Letters, 451, 136–141.
Katayama, M., Wiklander, O. P. B., Fritz, T., Caidahl, K., El-Andaloussi, S., Zierath, J. R., & Krook, A. (2019). Circulating exosomal miR-20b-5p is elevated in type 2 diabetes and could impair insulin action in human skeletal muscle. Diabetes, 68(3), 515–526.
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., Mirzaei, H., Goldsmith, E. J., Longgood, J., Pei, J., Grishin, N. V., Frantz, D. E., Schneider, J. W., Chen, S., Li, L., Sawaya, M. R., Eisenberg, D., Tycko, R., & McKnight, S. L. (2012). Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell, 149(4), 753–767.
Keppetipola, N., Sharma, S., Li, Q., & Black, D. L. (2012). Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Critical Reviews in Biochemistry and Molecular Biology, 47(4), 360–378.
Kim, T. H., Payliss, B. J., Nosella, M. L., Lee, I. T. W., Toyama, Y., Forman-Kay, J. D., & Kay, L. E. (2021). Interaction hot spots for phase separation revealed by NMR studies of a CAPRIN1 condensed phase. Proceedings of the National Academy of Sciences USA, 118(23), e2104897118.
Knowles, T. P., Vendruscolo, M., & Dobson, C. M. (2014). The amyloid state and its association with protein misfolding diseases. Nature Reviews Molecular Cell Biology, 15(6), 384–396.
Kretz, M., Siprashvili, Z., Chu, C., Webster, D. E., Zehnder, A., Qu, K., Lee, C. S., Flockhart, R. J., Groff, A. F., Chow, J., Johnston, D., Kim, G. E., Spitale, R. C., Flynn, R. A., Zheng, G. X., Aiyer, S., Raj, A., Rinn, J. L., Chang, H. Y., & Khavari, P. A. (2013). Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature, 493(7431), 231–235.
Kristensen, L. S., Jakobsen, T., Hager, H., & Kjems, J. (2022). The emerging roles of circRNAs in cancer and oncology. Nature Reviews. Clinical Oncology, 19(3), 188–206.
Kulkarni, M., Ozgur, S., & Stoecklin, G. (2010). On track with P-bodies. Biochemical Society Transactions, 38(Pt 1), 242–251.
Lallemand-Breitenbach, V., & de The, H. (2010). PML nuclear bodies. Cold Spring Harbor Perspectives in Biology, 2(5), a000661.
Larson, A. G., Elnatan, D., Keenen, M. M., Trnka, M. J., Johnston, J. B., Burlingame, A. L., Agard, D. A., Redding, S., & Narlikar, G. J. (2017). Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature, 547(7662), 236–240.
Lee, S., Kopp, F., Chang, T. C., Sataluri, A., Chen, B., Sivakumar, S., Yu, H., Xie, Y., & Mendell, J. T. (2016). Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell, 164(1–2), 69–80.
Leucci, E., Vendramin, R., Spinazzi, M., Laurette, P., Fiers, M., Wouters, J., Radaelli, E., Eyckerman, S., Leonelli, C., Vanderheyden, K., Rogiers, A., Hermans, E., Baatsen, P., Aerts, S., Amant, F., Van Aelst, S., van den Oord, J., de Strooper, B., Davidson, I., … Marine, J. C. (2016). Melanoma addiction to the long non-coding RNA SAMMSON. Nature, 531(7595), 518–522.
Li, C., Hu, J., Hu, X., Zhao, C., Mo, M., Zu, X., & Li, Y. (2021a). LncRNA SNHG9 is a prognostic biomarker and correlated with immune infiltrates in prostate cancer. Translational Andrology and Urology, 10(1), 215–226.
Li, R. H., Tian, T., Ge, Q. W., He, X. Y., Shi, C. Y., Li, J. H., Zhang, Z., Liu, F. Z., Sang, L. J., Yang, Z. Z., Liu, Y. Z., Xiong, Y., Yan, Q., Li, X., Ju, H. Q., Liu, J., Wang, L. J., Shao, J. Z., Wang, W., … Lin, A. (2021b). A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Research, 31(10), 1088–1105.
Li, L., Roy, K., Katyal, S., Sun, X., Bleoo, S., & Godbout, R. (2006). Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Molecular Biology of the Cell, 17(3), 1126–1140.
Li, X., Yang, L., & Chen, L. L. (2018). The biogenesis, functions, and challenges of circular RNAs. Molecular Cell, 71(3), 428–442.
Liu, C. X., & Chen, L. L. (2022). Circular RNAs: characterization, cellular roles, and applications. Cell, 185(12), 2016–2034.
Liu, J. L., & Gall, J. G. (2007). U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proceedings of the National Academy of Sciences USA, 104(28), 11655–11659.
Liu, J., Liu, Z. X., Wu, Q. N., Lu, Y. X., Wong, C. W., Miao, L., Wang, Y., Wang, Z., Jin, Y., He, M. M., Ren, C., Wang, D. S., Chen, D. L., Pu, H. Y., Feng, L., Li, B., Xie, D., Zeng, M. S., Huang, P., … Ju, H. Q. (2020). Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nature Communications, 11(1), 1507.
Liu, J., Rivas, F. V., Wohlschlegel, J., Yates, J. R., 3rd., Parker, R., & Hannon, G. J. (2005). A role for the P-body component GW182 in microRNA function. Nature Cell Biology, 7(12), 1261–1266.
Liu, X., He, B., Xu, T., Pan, Y., Hu, X., Chen, X., & Wang, S. (2018). MiR-490-3p functions as a tumor suppressor by inhibiting oncogene VDAC1 expression in colorectal cancer. Journal of Cancer, 9(7), 1218–1230.
Liu, X. M., Ma, L., & Schekman, R. (2021). Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. eLife, 10, e71982.
Lopez-Erauskin, J., Tadokoro, T., Baughn, M. W., Myers, B., McAlonis-Downes, M., Chillon-Marinas, C., Asiaban, J. N., Artates, J., Bui, A. T., Vetto, A. P., Lee, S. K., Le, A. V., Sun, Y., Jambeau, M., Boubaker, J., Swing, D., Qiu, J., Hicks, G. G., Ouyang, Z., … Da Cruz, S. (2018). ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron, 100(4), 816–830.
Luo, J., Qu, L., Gao, F., Lin, J., Liu, J., & Lin, A. (2021). LncRNAs: Architectural scaffolds or more potential roles in phase separation. Frontiers in Genetics, 12, 626234.
Ma, L., Bajic, V. B., & Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biology, 10(6), 925–933.
Maharana, S., Wang, J., Papadopoulos, D. K., Richter, D., Pozniakovsky, A., Poser, I., Bickle, M., Rizk, S., Guillén-Boixet, J., Franzmann, T. M., Jahnel, M., Marrone, L., Chang, Y. T., Sterneckert, J., Tomancak, P., Hyman, A. A., & Alberti, S. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science, 360(6391), 918–921.
Mannen, T., Yamashita, S., Tomita, K., Goshima, N., & Hirose, T. (2016). The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. Journal of Cell Biology, 214(1), 45–59.
Michelini, F., Pitchiaya, S., Vitelli, V., Sharma, S., Gioia, U., Pessina, F., Cabrini, M., Wang, Y., Capozzo, I., Iannelli, F., Matti, V., Francia, S., Shivashankar, G. V., Walter, N. G., & d’Adda di Fagagna, F. (2017). Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nature Cell Biology, 19(12), 1400–1411.
Modic, M., Grosch, M., Rot, G., Schirge, S., Lepko, T., Yamazaki, T., Lee, F. C. Y., Rusha, E., Shaposhnikov, D., Palo, M., Merl-Pham, J., Cacchiarelli, D., Rogelj, B., Hauck, S. M., von Mering, C., Meissner, A., Lickert, H., Hirose, T., Ule, J., & Drukker, M. (2019). Cross-regulation between TDP-43 and paraspeckles promotes pluripotency-differentiation transition. Molecular Cell, 74(5), 951–965. e913.
Mongelli, A., Martelli, F., Farsetti, A., & Gaetano, C. (2019). The dark that matters: long non-coding RNAs as master regulators of cellular metabolism in non-communicable diseases. Frontiers in Physiology, 10, 369.
Murray, D. T., Kato, M., Lin, Y., Thurber, K. R., Hung, I., McKnight, S. L., & Tycko, R. (2017). Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell, 171(3), 615–627. e616.
Naganuma, T., Nakagawa, S., Tanigawa, A., Sasaki, Y. F., Goshima, N., & Hirose, T. (2012). Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO Journal, 31(20), 4020–4034.
Ninomiya, K., Adachi, S., Natsume, T., Iwakiri, J., Terai, G., Asai, K., & Hirose, T. (2020). LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO Journal, 39(3), e102729.
Ninomiya, K., Iwakiri, J., Aly, M. K., Sakaguchi, Y., Adachi, S., Natsume, T., Terai, G., Asai, K., Suzuki, T., & Hirose, T. (2021). m(6) A modification of HSATIII lncRNAs regulates temperature-dependent splicing. EMBO Journal, 40(15), e107976.
Nizami, Z., Deryusheva, S., & Gall, J. G. (2010). The Cajal body and histone locus body. Cold Spring Harbor Perspectives in Biology, 2(7), a000653.
Noh, J. H., Kim, K. M., Abdelmohsen, K., Yoon, J. H., Panda, A. C., Munk, R., Kim, J., Curtis, J., Moad, C. A., Wohler, C. M., Indig, F. E., de Paula, W., Dudekula, D. B., De, S., Piao, Y., Yang, X., Martindale, J. L., de Cabo, R., & Gorospe, M. (2016). HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes & Development, 30(10), 1224–1239.
Noto, J. J., Schmidt, C. A., & Matera, A. G. (2017). Engineering and expressing circular RNAs via tRNA splicing. RNA Biology, 14(8), 978–984.
Nott, T. J., Petsalaki, E., Farber, P., Jervis, D., Fussner, E., Plochowietz, A., Craggs, T. D., Bazett-Jones, D. P., Pawson, T., Forman-Kay, J. D., & Baldwin, A. J. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Molecular Cell, 57(5), 936–947.
Onoguchi-Mizutani, R., & Akimitsu, N. (2022). Long noncoding RNA and phase separation in cellular stress response. Journal of Biochemistry, 171(3), 269–276.
Onoguchi-Mizutani, R., Kirikae, Y., Ogura, Y., Gutschner, T., Diederichs, S., & Akimitsu, N. (2021). Identification of a heat-inducible novel nuclear body containing the long noncoding RNA MALAT1. Journal of Cell Science, 134(10), jcs253559.
Ou, Y., He, J., & Liu, Y. (2018). MiR-490-3p inhibits autophagy via targeting ATG7 in hepatocellular carcinoma. IUBMB Life, 70(6), 468–478.
Pandya-Jones, A., Markaki, Y., Serizay, J., Chitiashvili, T., Mancia Leon, W. R., Damianov, A., Chronis, C., Papp, B., Chen, C. K., McKee, R., Wang, X. J., Chau, A., Sabri, S., Leonhardt, H., Zheng, S., Guttman, M., Black, D. L., & Plath, K. (2020). A protein assembly mediates Xist localization and gene silencing. Nature, 587(7832), 145–151.
Pessina, F., Giavazzi, F., Yin, Y., Gioia, U., Vitelli, V., Galbiati, A., Barozzi, S., Garre, M., Oldani, A., Flaus, A., Cerbino, R., Parazzoli, D., Rothenberg, E., & d’Adda di Fagagna, F. (2019). Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nature Cell Biology, 21(10), 1286–1299.
Pirrotta, V., & Li, H. B. (2012). A view of nuclear polycomb bodies. Current Opinion in Genetics & Development, 22(2), 101–109.
Qamar, S., Wang, G., Randle, S. J., Ruggeri, F. S., Varela, J. A., Lin, J. Q., Phillips, E. C., Miyashita, A., Williams, D., Ströhl, F., Meadows, W., Ferry, R., Dardov, V. J., Tartaglia, G. G., Farrer, L. A., Kaminski Schierle, G. S., Kaminski, C. F., Holt, C. E., Fraser, P. E., … St George-Hyslop, P. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell, 173(3), 720–734. e715.
Qin, D., Wei, R., Zhu, S., Min, L., & Zhang, S. (2021). MiR-490-3p silences CDK1 and inhibits the proliferation of colon cancer through an LLPS-dependent miRISC system. Frontiers in Molecular Biosciences, 8, 561678.
Qu, W., Wang, Z., & Zhang, H. (2020). Phase separation of the C. elegans Polycomb protein SOP-2 is modulated by RNA and sumoylation. Protein & Cell, 11(3), 202–207.
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., Patil, D. P., Kwak, H., Lee, J. H., & Jaffrey, S. R. (2019). m(6)A enhances the phase separation potential of mRNA. Nature, 571(7765), 424–428.
Saj, A., & Lai, E. C. (2011). Control of microRNA biogenesis and transcription by cell signaling pathways. Current Opinion in Genetics & Development, 21(4), 504–510.
Salzman, J. (2016). Circular RNA expression: its potential regulation and function. Trends in Genetics, 32(5), 309–316.
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., & Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE, 7(2), e30733.
Schertzer, M. D., Braceros, K. C. A., Starmer, J., Cherney, R. E., Lee, D. M., Salazar, G., Justice, M., Bischoff, S. R., Cowley, D. O., Ariel, P., Zylka, M. J., Dowen, J. M., Magnuson, T., & Calabrese, J. M. (2019). lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG island DNA. Molecular Cell, 75(3), 523–537. e510.
Schmitt, A. M., Garcia, J. T., Hung, T., Flynn, R. A., Shen, Y., Qu, K., Payumo, A. Y., Peres-da-Silva, A., Broz, D. K., Baum, R., Guo, S., Chen, J. K., Attardi, L. D., & Chang, H. Y. (2016). An inducible long noncoding RNA amplifies DNA damage signaling. Nature Genetics, 48(11), 1370–1376.
Sen, G. L., & Blau, H. M. (2005). Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nature Cell Biology, 7(6), 633–636.
Shan, Z., Tu, Y., Yang, Y., Liu, Z., Zeng, M., Xu, H., Long, J., Zhang, M., Cai, Y., & Wen, W. (2018). Basal condensation of Numb and Pon complex via phase transition during Drosophila neuroblast asymmetric division. Nature Communications, 9(1), 737.
Sheu-Gruttadauria, J., & MacRae, I. J. (2018). Phase transitions in the assembly and function of human miRISC. Cell, 173(4), 946–957. e916.
Shin, Y., & Brangwynne, C. P. (2017). Liquid phase condensation in cell physiology and disease. Science, 357(6357), eaaf4382.
Soghli, N., Yousefi, T., Abolghasemi, M., & Qujeq, D. (2021). NORAD, a critical long non-coding RNA in human cancers. Life Sciences, 264, 118665.
Spector, D. L., & Lamond, A. I. (2011). Nuclear speckles. Cold Spring Harbor Perspectives in Biology, 3(2), a000646.
Strom, A. R., & Brangwynne, C. P. (2019). The liquid nucleome—phase transitions in the nucleus at a glance. Journal of Cell Science, 132(22), jcs235093.
Su, M., Xiao, Y., Ma, J., Tang, Y., Tian, B., Zhang, Y., Li, X., Wu, Z., Yang, D., Zhou, Y., Wang, H., Liao, Q., & Wang, W. (2019). Circular RNAs in cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Molecular Cancer, 18(1), 90.
Su, Q., Mehta, S., & Zhang, J. (2021). Liquid-liquid phase separation: Orchestrating cell signaling through time and space. Molecular Cell, 81(20), 4137–4146.
Su, X., Ditlev, J. A., Hui, E., Xing, W., Banjade, S., Okrut, J., King, D. S., Taunton, J., Rosen, M. K., & Vale, R. D. (2016). Phase separation of signaling molecules promotes T cell receptor signal transduction. Science, 352(6285), 595–599.
Suk, T. R., & Rousseaux, M. W. C. (2020). The role of TDP-43 mislocalization in amyotrophic lateral sclerosis. Molecular Neurodegeneration, 15(1), 45.
Sun, X., Liu, J., Xu, C., Tang, S. C., & Ren, H. (2016). The insights of Let-7 miRNAs in oncogenesis and stem cell potency. Journal of Cellular and Molecular Medicine, 20(9), 1779–1788.
Suzuki, H., & Tsukahara, T. (2014). A view of pre-mRNA splicing from RNase R resistant RNAs. International Journal of Molecular Sciences, 15(6), 9331–9342.
Tichon, A., Gil, N., Lubelsky, Y., Havkin Solomon, T., Lemze, D., Itzkovitz, S., Stern-Ginossar, N., & Ulitsky, I. (2016). A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nature Communications, 7, 12209.
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., Freier, S. M., Bennett, C. F., Sharma, A., Bubulya, P. A., Blencowe, B. J., Prasanth, S. G., & Prasanth, K. V. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell, 39(6), 925–938.
Wang, B., Zhang, L., Dai, T., Qin, Z., Lu, H., Zhang, L., & Zhou, F. (2021a). Liquid-liquid phase separation in human health and diseases. Signal Transduction and Targeted Therapy, 6(1), 290.
Wang, R., Cao, L., Thorne, R. F., Zhang, X. D., Li, J., Shao, F., Zhang, L., & Wu, M. (2021b). LncRNA GIRGL drives CAPRIN1-mediated phase separation to suppress glutaminase-1 translation under glutamine deprivation. Science Advances, 7(13), eabe5708.
Wang, X., Liu, C., Zhang, S., Yan, H., Zhang, L., Jiang, A., Liu, Y., Feng, Y., Li, D., Guo, Y., Hu, X., Lin, Y., Bu, P., & Li, D. (2021c). N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Developmental Cell, 56(5), 702–715. e708.
Wang, C., Duan, Y., Duan, G., Wang, Q., Zhang, K., Deng, X., Qian, B., Gu, J., Ma, Z., Zhang, S., Guo, L., Liu, C., & Fang, Y. (2020a). Stress induces dynamic, cytotoxicity-antagonizing TDP-43 nuclear bodies via paraspeckle LncRNA NEAT1-mediated liquid-liquid phase separation. Molecular Cell, 79(3), 443-458.e447.
Wang, D., Cao, X., Han, Y., & Yu, D. (2020b). LncRNA SNHG9 is downregulated in non-small cell lung cancer and suppressed miR-21 through methylation to promote cell proliferation. Cancer Manag Res, 12, 7941–7948.
Wang, M., Tao, X., Jacob, M. D., Bennett, C. A., Ho, J. J. D., Gonzalgo, M. L., Audas, T. E., & Lee, S. (2018). Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Reports, 24(7), 1713-1721.e1714.
Woodruff, J. B., Hyman, A. A., & Boke, E. (2018). Organization and function of non-dynamic biomolecular condensates. Trends in Biochemical Sciences, 43(2), 81–94.
Wu, H., Yin, Q. F., Luo, Z., Yao, R. W., Zheng, C. C., Zhang, J., Xiang, J. F., Yang, L., & Chen, L. L. (2016). Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Molecular Cell, 64(3), 534–548.
Xing, Z., Zhang, Y., Liang, K., Yan, L., Xiang, Y., Li, C., Hu, Q., Jin, F., Putluri, V., Putluri, N., Coarfa, C., Sreekumar, A., Park, P. K., Nguyen, T. K., Wang, S., Zhou, J., Zhou, Y., Marks, J. R., Hawke, D. H., … Lin, C. (2018). Expression of long noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer Research, 78(16), 4524–4532.
Yamazaki, T., Souquere, S., Chujo, T., Kobelke, S., Chong, Y. S., Fox, A. H., Bond, C. S., Nakagawa, S., Pierron, G., & Hirose, T. (2018). Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Molecular Cell, 70(6), 1038-1053.e1037.
Yang, P., Mathieu, C., Kolaitis, R. M., Zhang, P., Messing, J., Yurtsever, U., Yang, Z., Wu, J., Li, Y., Pan, Q., Yu, J., Martin, E. W., Mittag, T., Kim, H. J., & Taylor, J. P. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell, 181(2), 325–345. e328.
Yap, K., Mukhina, S., Zhang, G., Tan, J. S. C., Ong, H. S., & Makeyev, E. V. (2018). A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Molecular Cell, 72(3), 525-540.e513.
Yin, Q. F., Yang, L., Zhang, Y., Xiang, J. F., Wu, Y. W., Carmichael, G. G., & Chen, L. L. (2012). Long noncoding RNAs with snoRNA ends. Molecular Cell, 48(2), 219–230.
Yoo, H., Triandafillou, C., & Drummond, D. A. (2019). Cellular sensing by phase separation: Using the process, not just the products. Journal of Biological Chemistry, 294(18), 7151–7159.
Zeng, M., Chen, X., Guan, D., Xu, J., Wu, H., Tong, P., & Zhang, M. (2018). Reconstituted postsynaptic density as a molecular platform for understanding synapse formation and plasticity. Cell, 174(5), 1172–1187. e1116.
Zeng, M., Shang, Y., Araki, Y., Guo, T., Huganir, R. L., & Zhang, M. (2016). Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell, 166(5), 1163–1175. e1112.
Zhang, P., Wu, W., Chen, Q., & Chen, M. (2019a). Non-coding rnas and their integrated networks. Journal of integrative Bioinformatics, 16(3), 1–12.
Zhang, X. T., Pan, S. X., Wang, A. H., Kong, Q. Y., Jiang, K. T., & Yu, Z. B. (2019b). Long non-coding RNA (lncRNA) X-Inactive specific transcript (XIST) plays a critical role in predicting clinical prognosis and progression of colorectal cancer. Medical Science Monitor, 25, 6429–6435.
Zhao, Y. G., & Zhang, H. (2020). Phase separation in membrane biology: The interplay between membrane-bound organelles and membraneless condensates. Developmental Cell, 55(1), 30–44.
Zogg, H., Singh, R., & Ro, S. (2022). Current advances in RNA therapeutics for human diseases. International Journal of Molecular Sciences, 23(5), 2736.
Acknowledgements
This work was supported by funding from National Natural Science Foundation of China under Grant 81820108021 and U2004138.
Author information
Authors and Affiliations
Translational Research Institute, Henan Provincial People’s Hospital, Academy of Medical Sciences, Zhengzhou University, Zhengzhou, 450053, Henan, China
Mingyue Li, Rick F. Thorne, Xu Dong Zhang, Mian Wu & Song Chen
Corresponding authors
Correspondence to Mian Wu or Song Chen.
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, M., Thorne, R.F., Zhang, X.D. et al. Drops in the cell ocean: new roles for non-coding RNAs in liquid–liquid phase separation. GENOME INSTAB. DIS. (2022). https://doi.org/10.1007/s42764-022-00091-0
Received21 August 2022
Revised28 October 2022
Accepted31 October 2022
Published19 November 2022
DOIhttps://doi.org/10.1007/s42764-022-00091-0
Share this article
Anyone you share the following link with will be able to read this content:
Keywords
Long non-coding RNA
microRNA
Circular RNA
Liquid–liquid phase separation
Cancer








用户登录
还没有账号?
立即注册