PDHE1α in DNA damage repair: a critical chromatin acetylation regulator
Commentary
Published: 11 November 2023
Bingsong Huang, Yuping Chen & Jian Yuan, Volume 4, pages 349–350, (2023)
Abstract
A recent study in Nature Structural and Molecular Biology reveals that chromatin-associated PDHE1α generates acetyl-CoA near DNA double-strand breaks, crucial for chromatin remodeling and DNA repair. PDHE1α recruitment depends on PARP1 and impacts genome stability and cancer therapy resistance. This research sheds light on DNA damage response and chromatin acetylation regulation.
Local chromatin acetylation around DSBs is essential for the relaxation of chromatin structure following DNA damage (Murr et al., 2006; Sivanand et al., 2017; Shogren-Knaak et al., 2006; Xu et al., 2012), which is a precondition for the initiation of double-strand break (DSB) repair. However, a remaining question is how a substantial amount of acetyl-coenzyme A (acetyl-CoA), the precursor for acetylation reactions, is generated after DNA damage. A large amount of acetyl-CoA can be produced in mitochondria, however prior studies have shown that mitochondrial dysfunction does not impact the levels of nuclear acetyl-CoA (Yi et al., 2011), implying the existence of an mitochondrial independent regulatory mechanism for nuclear acetyl-CoA generation, especially in DNA damage response (Fig. 1).
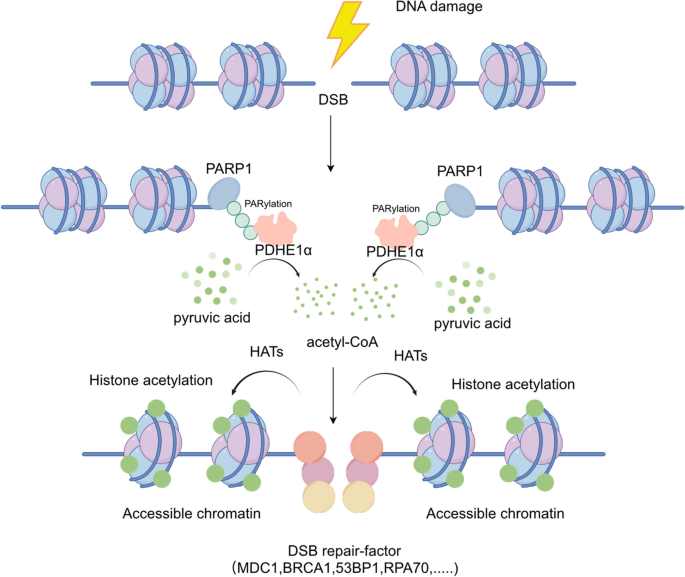
After DNA damage, PDHE1α is recruited to double-strand break (DSB) sites in a PARylation-dependent manner mediated by PARP1. Within the chromatin environment, PDHE1α catalyzes pyruvate metabolism, leading to the localized accumulation of acetyl-CoA near DSB sites. This localized accumulation plays a pivotal role in promoting chromatin relaxation, which, in turn, enhances DSB repair-factor recruitment and subsequent DNA repair. HATs, histone acetyltransferases
In a recent publication in Nature Structural & Molecular Biology (Zhang et al., 2023), Jun Zhang and the colleagues reveals the mechanism by which chromatin-associated pyruvate dehydrogenase E1 alpha (PDHE1α) locally generates acetyl-CoA to reshape the chromatin environment near DSBs and facilitate their repair. This study provides a long-awaited solution to the puzzle of the origin of nuclear acetyl-CoA. In the study, they employed AsiSI-induced DNA double-strand breaks (DSBs) in cell lines and conducted Chromatin Immunoprecipitation (ChIP) assays with pan-acetylation-specific antibodies. The results revealed a notable enrichment of acetylation levels at the DSB sites, with a peak in nuclear acetylation levels observed 1–2 h post-DNA damage induction. Additionally, the study demonstrated the recruitment of pyruvate dehydrogenase E1 alpha (PDHE1α) to the DNA damage sites following damage. Laser stripe experiments further confirmed the co-localization of PDHE1α with γH2AX and pan-acetylation signals. Subsequently, the researchers identified a rapid accumulation of pyruvate at the damage sites post-damage. Through isotope labeling and mass spectrometry, they unveiled a range of acetylated proteins in proximity to DNA damage sites. These findings illustrated the interaction between chromatin remodeling, acetyl-CoA metabolism, and the DNA damage response (DDR).
The authors further explored into the mechanisms governing the recruitment of PDHE1α to DSBs. Mass spectrometry analysis revealed the interaction between PARP1 and PDHE1α. Inhibiting PARP activity or reducing PARP1 levels effectively hindered the recruitment of PDHE1α to chromatin, indicating the pivotal role of PARP1 in PDHE1α recruitment. Furthermore, PARP1 was shown to PARylate PDHE1α, with critical binding regions identified in the 91–190 domain, including key binding sites at C91/94, H113/121, and modification sites at D95/E98/T111/D112/T124/T126. Notably, PDHE1α PARylation was found to influence chromatin accessibility. Intriguingly, although PDHE1α's enzymatic activity did not affect its recruitment to chromatin, it still had an impact on chromatin accessibility. This highlights the significance of PDHE1α in generating acetyl-CoA, which, in turn, affects chromatin accessibility.
In addition, the author uncovered that PDHE1α influences both Homologous Recombination (HR) and Non-Homologous End Joining (NHEJ) repair pathways, thus impacting genome stability. Clinical samples and animal experiments corroborated the link between PDHE1α and tumor resistance to DNA damage therapy, particularly through its PARylation activity,
In conclusion, the study identified PDHE1α as a crucial metabolic enzyme responsible for generating acetyl-CoA within the nucleus. Its recruitment to DNA damage sites depends on PARylation and facilitates local protein acetylation, consequently influencing chromatin states, particularly chromatin accessibility. This study shed light on the intricate processes involved in DNA damage repair and chromatin acetylation regulation. Understanding the role of PDHE1α in producing acetyl-CoA within the nucleus paves the way for potential applications in enhancing cancer treatment and preserving genome stability. Taken together, This study contributes to the understanding of the DNA damage response and opens up new possibilities for therapeutic interventions in cancer treatment.
References
Murr, R., et al. (2006). Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature Cell Biology, 8, 91–99. https://doi.org/10.1038/ncb1343
Shogren-Knaak, M., et al. (2006). Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science, 311, 844–847. https://doi.org/10.1126/science.1124000
Sivanand, S., et al. (2017). Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Molecular Cell, 67, 252-265.e256. https://doi.org/10.1016/j.molcel.2017.06.008
Xu, Y., et al. (2012). Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Molecular Cell, 48, 723–733. https://doi.org/10.1016/j.molcel.2012.09.026
Yi, C. H., et al. (2011). Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell, 146, 607–620. https://doi.org/10.1016/j.cell.2011.06.050
Zhang, J., et al. (2023). PARylated PDHE1α generates acetyl-CoA for local chromatin acetylation and DNA damage repair. Nature Structural & Molecular Biology. https://doi.org/10.1038/s41594-023-01107-3
Author information
Authors and Affiliations
Department of Biochemistry and Molecular Biology, Tongji University School of Medicine, Shanghai, 200120, China
Bingsong Huang, Yuping Chen & Jian Yuan
Department of Neurosurgery, Shanghai East Hospital, Tongji University School of Medicine, 150 Jimo Road, Shanghai, 200120, China
Bingsong Huang
Contributions
JY designed the study. BH wrote the manuscript. YC reviewed and edited the manuscript.
Corresponding author
Correspondence to Jian Yuan.
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in relation to this work.
Rights and permissions
About this article
Cite this article
Huang, B., Chen, Y. & Yuan, J. PDHE1α in DNA damage repair: a critical chromatin acetylation regulator. GENOME INSTAB. DIS. 4, 349–350 (2023). https://doi.org/10.1007/s42764-023-00112-6
Received15 October 2023
Revised15 October 2023
Accepted17 October 2023
Published11 November 2023
Issue DateDecember 2023
DOIhttps://doi.org/10.1007/s42764-023-00112-6
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable link







用户登录
还没有账号?
立即注册