H3K9me3 asymmetry: epigenetic choreography in DNA replication for genomic stability
Commentary
Published: 28 November 2023
Wei-Guo Zhu
Genome Instability & Disease Volume 4, pages 351–353, (2023)
Abstract
Parental histones, which are modified distinctively from their newly synthesized counterparts, are recycled during DNA replication for the re-establishment of a functional epigenome in the daughter cells. However, the mechanisms and functional implications underlying parental histone deposition onto replicating DNA strands remain enigmatic. A recent study published in Nature reveals a unique pattern of H3K9me3 distribution during DNA replication, which is governed by the human silencing hub (HUSH) complex and DNA polymerase Pol ε. H3K9me3 asymmetry toward the leading strand is important for the silencing of L1 retrotransposons, thus safeguarding both the epigenomic and genomic integrity.
Main
Histones are fundamental structural proteins that constitute the basic units of eukaryotic chromatin, known as nucleosomes. Their post-translational modifications serve as one of the crucial pillars of the epigenetic machinery, which play key regulatory roles in biological processes, such as chromatin structure organization, gene expression, and DNA damage repair (Bannister & Kouzarides, 2011). Parental histones covalently modified by specific marks represent essential carriers of epigenetic information, which needs to be faithfully restored to maintain the transcription programs and cell identify (Escobar et al., 2021; Stewart-Morgan et al., 2020). However, the precise mechanisms governing their accurate transmission to progeny cells during cell division and DNA replication remain poorly understood. Furthermore, the diverse array of over a hundred types of identified histone modifications raises questions about whether they follow similar distribution patterns.
H3K9me3, as the most crucial repressive histone mark in higher eukaryotic cells, plays a vital role in suppressing the widespread repetitive sequences within the genome and maintaining genomic stability. Nevertheless, the pattern and mechanism underlying its distribution during DNA replication remain unclear. Recently, Li and colleagues from Columbia University, building upon their previous work, published a comprehensive research article titled “Asymmetric distribution of parental H3K9me3 in S phase silences L1 elements” in Nature (Li et al., 2023). This groundbreaking study unveils, for the first time, the distribution mechanism and biological significance of H3K9me3 during DNA replication.
Early studies have hypothesized that parental histones are randomly and evenly distributed to newly synthesized daughter strands, namely the leading and lagging strands, during DNA replication (Leffak et al., 1977; Seale, 1976). Subsequently, they form nascent nucleosomes together with newly synthesized histones, assembling into progeny chromatin to ensure the accurate transmission of epigenetic information between generations. However, due to technological limitations, this hypothesis remained unverified for many years, and the molecular mechanisms involved were largely obscure until very recently. Particularly, several replisome proteins have been identified to possess histone chaperone activity and aid in parental histone partitioning during DNA replication. For example, MCM2 and Pol α, both carrying a conserved N-terminal histone-binding motif, facilitate the transfer of parental histone onto the lagging strands (Gan et al., 2018; Li et al., 2020; Petryk et al., 2018). On the other hand, POLE3/POLE4 (Dpb4/Dpb3 in yeast), the accessory subunits of the leading strand DNA polymerase Pol ε, promote parental histone deposition onto the leading strands (Li et al., 2020; Yu et al., 2018). To further investigate this process in mammalian cells, the authors improved the eSPAN pipeline, originally developed in budding yeast (Yu et al., 2014), by combining the CUT&Tag and BrdU immunoprecipitation, which allows for efficient and strand-specific profiling of proteins of interest on nascent chromatin (Li et al., 2020, 2021). Using this revamped eSPAN technique, Li et al. screened the distribution patterns of about a dozen commonly studied histone modifications in mouse embryonic stem cells (Li et al., 2023). While the majority of histone marks were distributed in an almost symmetrical manner between the two newly synthesized strands, H3K9me3 displayed a surprisingly strong and unique preference toward the leading strand.
Subsequently, the authors delved deeper into this phenomenon. Through bioinformatic analysis, they found that the asymmetric distribution of H3K9me3 primarily occurs at long interspersed nuclear elements (LINE or L1), especially in L1 sequences with transcriptional orientation head-on to the movement of DNA replication forks. L1, as the most abundant transposable element, constitutes approximately 17% of the human genome and is widely distributed in various regions, including both euchromatin and heterochromatin (Lander et al., 2001). Further analysis revealed that L1 sequences affected by the asymmetric distribution of H3K9me3 are evolutionarily younger and longer. Previous reports had indicated that these L1 elements are regulated by the human silencing hub (HUSH) complex (Liu et al., 2018; Tchasovnikarova et al., 2015). Indeed, the authors confirmed that the asymmetry of H3K9me3 distribution is also regulated by the HUSH complex by knocking out different HUSH subunits.
However, depletion of HUSH subunits only partially reduced H3K9me3 asymmetry, and it could not explain its unique preference for the leading strand. To address this, the authors further explored other potential regulatory factors. As mentioned above, the previous studies suggested that the leading strand DNA polymerase Pol ε is involved in the transfer of parental histones to the leading strand (Li et al., 2020; Yu et al., 2018). By knocking out its non-essential subunits POLE3/4, the authors confirmed that Pol ε also participates in the deposition of H3K9me3. Intuitively, the authors proposed a mutual interaction between Pol ε and HUSH that coordinates the asymmetric distribution of H3K9me3. Immunoprecipitation and in vitro-binding assays confirmed a direct binding between Pol ε and the HUSH complex, and mutations in the interacting region significantly reduced H3K9me3 asymmetry. These data suggest that the asymmetric distribution of H3K9me3 is jointly regulated by the HUSH complex and Pol ε, with both interacting to ensure the specificity for L1 and the bias toward the leading strand during DNA replication.
The asymmetry of H3K9me3 distribution dynamically changes with the progression of the cell cycle, reaching its maximum during the S phase, whereas L1 expression undergoes an S-phase-specific silencing, suggesting that H3K9me3 asymmetry plays an inhibitory role of L1 activation during DNA replication (Fig. 1). Indeed, the authors analyzed all the mutant cells showing compromised H3K9me3 asymmetry and found varying degrees of increased L1 expression. In normal cells, L1 is highly suppressed, because its abnormal expression is often associated with L1 retrotransposition and genomic instability. However, in cells with POLE4 or MPP8 knockout, the degree of L1 activation and the decrease in H3K9me3 asymmetry were significantly correlated. Analysis of publicly available datasets revealed a high concordance between L1 insertion into the genome and the leading strand bias of H3K9me3 distribution. Using an L1 retrotransposition reporter system, the authors confirmed that mutants defective of asymmetric H3K9me3 distribution increased L1 retrotransposition to different levels, ultimately leading to genomic instability. These results indicate that the asymmetry of H3K9me3 distribution effectively suppresses L1 expression and retrotransposition during DNA replication, thus maintaining genomic stability.
Fig. 1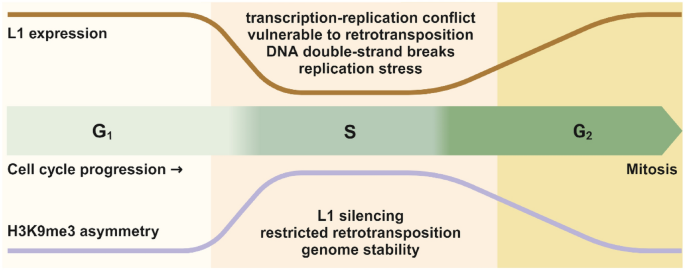
The dynamic choreography of H3K9me3 asymmetry and L1 expression during cell cycle progression. During S phase, the chromatin adopts a more accessible formation, which allows for efficient DNA synthesis, but also renders it more vulnerable to genotoxic stress. Aberrant L1 expression during S phase could lead to retrotransposition, potential transcription–replication conflict, replication stress, and even DNA double-strand breaks. H3K9me3 asymmetry thus serves as a safeguarding mechanism to prevent these detrimental effects and preserve genome stability
In summary, this study reveals the distribution mechanism of H3K9me3 during DNA replication and a novel regulatory paradigm for L1 silencing, which likely depicts a temporary response of cells to cope with the dilution of H3K9me3 by newly synthesized histones during DNA replication, maximizing the inhibitory function of H3K9me3 to protect the chromatin during the inherently vulnerable replication period. However, this model still raises some questions that warrant further exploration in future studies. For example, how is H3K9me3 asymmetry resolved outside of S phase and what is the role of SETDB1 or other methyltransferases in this process? How does H3K9me3 asymmetry toward the leading strand inhibit L1 expression? Answers to these questions will provide more insights into this critical yet little known process and advance our understandings of epigenetic inheritance.
Data availability
Not applicable.
References
Bannister, A. J., & Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Research, 21, 381–395.
Escobar, T. M., Loyola, A., & Reinberg, D. (2021). Parental nucleosome segregation and the inheritance of cellular identity. Nature Reviews Genetics, 22, 379–392.
Gan, H. Y., et al. (2018). The Mcm2-Ctf4-Pol alpha axis facilitates parental histone H3–H4 transfer to lagging strands. Molecular Cell, 72, 140–151.
Lander, E. S., et al. (2001). Initial sequencing and analysis of the human genome. Nature, 409, 860–921.
Leffak, I. M., Grainger, R., & Weintraub, H. (1977). Conservative assembly and segregation of nucleosomal histones. Cell, 12, 837–845.
Li, Z., Hua, X., Serra-Cardona, A., Xu, X., & Zhang, Z. (2021). Efficient and strand-specific profiling of replicating chromatin with enrichment and sequencing of protein-associated nascent DNA in mammalian cells. Nature Protocols, 16, 2698–2721.
Li, Z., et al. (2023). Asymmetric distribution of parental H3K9me3 in S phase silences L1 elements. Nature, 623, 643–651.
Li, Z. M., et al. (2020). DNA polymerase alpha interacts with H3–H4 and facilitates the transfer of parental histones to lagging strands. Science Advances. https://doi.org/10.1126/sciadv.abb5820
Liu, N., et al. (2018). Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature, 553, 228–232.
Petryk, N., et al. (2018). MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science, 361, 1389–1391.
Seale, R. L. (1976). Studies on the mode of segregation of histone nu bodies during replication in HeLa cells. Cell, 9, 423–429.
Stewart-Morgan, K. R., Petryk, N., & Groth, A. (2020). Chromatin replication and epigenetic cell memory. Nature Cell Biology, 22, 361–371.
Tchasovnikarova, I. A., et al. (2015). GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science, 348, 1481–1485.
Yu, C., et al. (2018). A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science, 361, 1386–1389.
Yu, C. H., et al. (2014). Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Molecular Cell, 56, 609–609.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32090030, 81720108027 and 81530074 to W.-G.Z); the National Key R&D Program of China (Grant No. 2017YFA0503900 to W.-G.Z.); the Science and Technology Program of Guangdong Province in China (Grant No. 2017B030301016 to W.-G.Z.); the Shenzhen Municipal Commission of Science and Technology Innovation (Grant Nos. JCYJ20200109114214463 and JCYJ20220818100015032 to W.-G.Z.); and Shenzhen University 2035 Program for Excellent Research to W.-G.Z.
Author information
Authors and Affiliations
Guangdong Key Laboratory for Genome Stability and Human Disease Prevention, Department of Biochemistry and Molecular Biology, Shenzhen University Medical School, Shenzhen, 518055, China
Wei-Guo Zhu
Shenzhen Bay Laboratory, Shenzhen University Medical School, Shenzhen, 518055, China
Wei-Guo Zhu
International Cancer Center, Shenzhen University Medical School, Shenzhen, 518055, China
Wei-Guo Zhu
Marshall Laboratory of Biomedical Engineering, Shenzhen University Medical School, Shenzhen, 518055, China
Wei-Guo Zhu
Corresponding author
Correspondence to Wei-Guo Zhu.
Ethics declarations
Conflict of interest
Wei-Guo Zhu is the Editor-in-Chief of Genome Instability & Disease.
Rights and permissions
About this article
Cite this article
Zhu, WG. H3K9me3 asymmetry: epigenetic choreography in DNA replication for genomic stability. GENOME INSTAB. DIS. 4, 351–353 (2023). https://doi.org/10.1007/s42764-023-00117-1
Received17 November 2023
Revised17 November 2023
Accepted20 November 2023
Published28 November 2023
Issue DateDecember 2023
DOIhttps://doi.org/10.1007/s42764-023-00117-1
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable link







用户登录
还没有账号?
立即注册