Death-associated protein 3 in cell death and beyond
Review Article
Published: 04 March 2024
Ting Cao, Xuling Luo, Binjiao Zheng, Yao Deng, Yu Zhang, Yuyan Li, Wenwen Xi, Meng Guo, Xuefeng Yang, Zhiyue Li & Bin Lu
Genome Instability & Disease Volume 5, (2024)
Abstract
Death-associated protein 3 (DAP3) is a highly conserved guanosine triphosphate (GTP) binding protein. As a component of the mitochondrial ribosome 28 S small subunit, DAP3 is involved in apoptosis pathways and plays a vital role in mitochondrial dynamics, mitochondrial protein synthesis, anoikis, and autophagy. Recently, DAP3 has been reported to participate in the development and progression of various cancers. In this review, we provide a brief overview of recent findings regarding the structure, subcellular localization, and function of DAP3 as well as its role in cancer development and progression.
Introduction
Death-associated protein 3 (DAP3), also known as mitochondrial ribosomal protein S29 (MRP S29), is encoded by a nuclear gene located on human chromosome 1q21, with a corresponding mRNA length of 1.7 kb (Kissil & Kimchi, 1997). The DAP3 protein consists of 398 amino acids, with an approximate molecular weight of 46 kDa. This highly conserved protein has guanosine triphosphate (GTP) binding activity (Kissil et al., 1995).Mitochondria serve as the primary site for aerobic respiration in eukaryotes and not only play a crucial role in biological energy metabolism and biosynthesis, but are important for regulation of related signaling pathways. As a component of the mitochondrial ribosome 28 S small subunit (Morgan et al., 2001; Suzuki et al., 2001), DAP3 participates in the synthesis of mitochondrial encoded proteins (Xiao et al., 2015; Kim et al., 2007) and influences the cell cycle by modulating mitochondrial dynamics, apoptosis, anoikis, and autophagy. In recent years, an increasing number of studies have revealed a close association between DAP3 and the occurrence and progression of various tumors. In this review, we provide a brief overview of recent findings regarding the structure, subcellular localization, and functions of DAP3 in mitochondrial dynamics, mitochondrial protein synthesis, anoikis, and autophagy as well as its role in cancer development and progression. We also consider the roles of DAP3 in health and disease and the potential for DAP3 as a diagnostic, prognostic and therapeutic target.
DAP3 structure
The structure of DAP3 exhibits a high degree of evolutionary conservation, with mouse and human DAP3 showing greater than 80% amino acid homology (Cavdar Koc et al., 2001). In addition, 35% of the amino acids of nematode DAP3 are identical to those of human DAP3, while 64% are similar. However, human DAP3 contains 13 exons, while nematode DAP3 has only seven (Morgan et al., 2001; Kissil et al., 1999). The structure of DAP3 consists primarily of three components (Fig. 1): the mitochondrial target sequence (MTS), phosphate-binding loop (P-loop), and nuclear receptor (NR).
Fig. 1
The domain structure and functional motifs within the primary amino acid sequence of human mitochondrial ribosome protein DAP3. MTS, mitochondrial target sequence; NR, nuclear receptor-interacting domain; P-loop, phosphate binding loop
The MTS is a peptide that locates proteins in the mitochondrial matrix. This signal peptide exists at the N-terminus of the protein sequence and is composed of amphoteric amino acids (having both hydrophilic and hydrophobic properties), several hydrophobic amino acids and charged amino acids (Morgan et al., 2001). The N-terminal signal peptide is cleaved when the DAP3 precursor enters the mitochondria. In human DAP3, the N-terminal cleavage site is located at amino acid position 18, while the site is located is located at amino acid position 17 in mouse DAP3 (Morgan et al., 2001).
The P-loop domain is critical for the pro-apoptotic function of DAP3. While DAP3 promotes apoptosis in 293T cells when the P-loop region remains intact, apoptosis is inhibited by deleting the first 100 amino acids at the N-terminal. Mutation of lysine at position 143 in the P-loop domain to alanine (K143A) noticeably also inhibited the apoptosis-promoting function of DAP3. In addition, ∆230 mutant (a mutant that lacks 168 C-terminal amino acids) of DAP3 significantly reduced cell death. Interestingly, the impactful negative effects of the ∆230 mutant were entirely contingent upon the existence of a functional P-loop motif ( Kissil et al., 1999). The P-loop domain serves as a phosphate-binding domain, indicating that DAP3 functions as an ATP/GTP-binding protein. Ultraviolet crosslinking of purified DAP3 in vitro revealed that DAP3 activates pro-caspase 8 in the presence of GTP, while this process is inhibited by GTPγS. However, no marked differences were observed in the effects of ATP and ATPγS on the activation of pro-caspase8 by DAP3, leading to the speculation that the P-loop domain of DAP3 binds to GTP rather than ATP (Miyazaki & Reed, 2001).
Located near the P-loop domain, the NR domain may be the site at which DAP3 binds to nuclear receptors such as glucocorticoid receptors (GRs), thyroid hormone receptors, and peroxisome proliferator-activated receptors (Morgan et al., 2001). Hulkko et al. reported that human mitochondrial ribosomes bind with high affinity to GDP and GTP in a 1:1 ratio (Hulkko & Zilliacus, 2002). However, no other mitochondrial ribosomal proteins with GTP-binding function have been identified; therefore, DAP3 may be an important medium by which GTP binds to human mitochondrial ribosomes. Mitochondrial ribosome protein S27 (MRP S27), another protein in the mitochondrial ribosome subunit, has guanine nucleotide exchange function and acts as a functional chaperone for DAP3 (Cavdar Koc et al., 2001). DAP3 contains three potential GTP-binding regions, GXXXXGK(S/T), DXXG, and NKXE, located at amino acid positions 128–135, 238–241, and 211–214, respectively (Cavdar Koc et al., 2001). In addition to these GTP-binding phosphorylation sites, DAP3 is also predicted to contain cAMP- and cGMP-dependent protease phosphorylation sites, which may play an important role in the mechanism by which DAP3 promotes apoptosis (Cavdar Koc et al., 2001). Miller et al. discovered additional phosphorylation sites in DAP3 at Ser215, Thr216, Ser220, Ser251, Ser252 and Ser280, which are located adjacent to the GTP binding domain, suggesting that they may serve as regulatory sites for DAP3 functionality (Miller et al., 2008). Thus, the domain structure of DAP3 underpins its functions as a GTP-binding mitoribosomal protein involved in a variety of processes including cellular respiration and cell death.
Subcellular localization of DAP3
MITOPROT and Target P software predict that human DAP3 is localized predominantly within mitochondria. However, the subcellular localization of DAP3 varies in different cells or under different physiological and pathological conditions (Morgan et al., 2001) and is crucial for its function. Although DAP3 contains an MTS in its N-terminal domain, there are currently two theories regarding its subcellular localization. First, DAP3 resides primarily in mitochondria as a component of mitochondrial ribosome subunits, with minimal presence in the cytoplasm and second, a cytoplasmic pool of DAP3 may exist in addition to its mitochondrial localization (Morgan et al., 2001).
Based on immunofluorescence studies, Morgan et al. demonstrated that DAP3 is localized in the human fibroblasts mitochondria (Morgan et al., 2001). In addition, Mukamel and Kimchi (Mukamel & Kimchi, 2004) detected DAP3 in the mitochondria of the HeLa and A549 tumor cell lines, as well as human and rodent fibroblasts. Furthermore, Xiao et al. observed DAP3 in the mitochondria rather than in the cytoplasm of HeLa, HEK293T (human embryonic kidney cells), and mouse embryonic fibroblast cell lines, with predominant localization in the mitochondrial matrix (Xiao et al., 2015). DAP3 interacts with heat shock protein 70 (HSP70) and this interaction is prevented when 17 amino acids in the N-terminal domain are removed due to the loss of the mitochondrial localization signal. Consequently, the mutant form of DAP3 is distributed in the cytoplasm. DAP3 interacts with glucocorticoid receptors (GR) or other nuclear receptors (Hulkko et al., 2000) via the N-terminal region to stabilize the function of GR in apoptosis by regulating its activity and the HSP90 complex in the cytoplasm (Hulkko & Zilliacus, 2002). These findings suggest that HSP70 functions as a companion molecule that allows DAP3 to enter mitochondria.
Although immunohistochemical staining of thyroid cancer tissue has shown that the majority of DAP3 is localized in mitochondria (Jacques et al., 2009), Sanna et al. reported that DAP3 is also present in the nucleus of COS-7 (fibroblast-like) cell lines (Hulkko & Zilliacus, 2002). Furthermore, DAP3 is associated with tumor necrosis factor (TNF) family death receptor complexes in the cytoplasm, especially FAS-associated death domain protein (FADD) (Miyazaki & Reed, 2001; Berger & Kretzler, 2002).
Thus, although DAP3 is distributed mainly in the mitochondrial matrix (Hulkko & Zilliacus, 2002), the localization of DAP3 in the cytoplasm and nucleus may be cell-specific and influenced by the physiological state of the cell. Therefore, further research is necessary to gain a more in-depth understanding of the subcellular localization of DAP3.
DAP3 and mitochondrial dynamics
Mitochondria consist of the inner and outer membranes, an intermembrane space, and the matrix. The shape and structure of mitochondria change constantly through fission and fusion of the inner and outer membranes. These two opposing processes represent a dynamic interplay that is essential for mitochondrial function (Lee et al., 2023; Yepuri et al., 2023). DAP3 maintains homeostasis in mitochondrial fusion and division by regulating phosphorylation of dynamin-related protein 1 at Ser637 (Xiao et al., 2015). DAP3 has been shown to participate in the process of mitochondrial division. In HeLa cells and CHO cells, exogenous labeled luciferase studies revealed that the morphology of mitochondria is long and tubular. However, following DAP3 overexpression, mitochondrial morphology becomes punctate, fragmented and concentrated around the nucleus, indicating that DAP3 overexpression promotes mitochondrial fission (Mukamel & Kimchi, 2004). Under stress conditions, DAP3 overexpression induces changes in mitochondrial structure accompanied by increased mitochondrial division and loss of mitochondrial membrane potential, which cause apoptosis. In contrast, reduced expression of DAP3 in HeLa cells significantly increased mitochondrial fragmentation (Xiao et al., 2015). Overexpression of DAP3 restored the tubular shape of fragmented mitochondria although, it should be noted that DAP3 overexpression alone did not alter mitochondrial morphology. However, these two statements about the relationship between DAP3 and mitochondrial dynamics are diametrically opposed. The former suggests that exogenous DAP3 overexpression leads to increased mitochondrial division, while reducing DAP3 expression requires stimulation by apoptotic drugs to decrease mitochondrial division. The latter conclusion is based on the proposition that reducing endogenous DAP3 expression increases mitochondrial division; however, exogenous overexpression of DAP3 alone does not affect cell division. Therefore, the precise role of DAP3 in mitochondrial fusion and division and the underlying mechanisms require further investigation.
DAP3 regulates mitochondrial protein synthesis
Mammalian mitochondria are semi-autonomous organelles with independent mitochondrial DNA (mtDNA), and translational machinery such as ribosomes, which replicate, transcribe, and translate independently of the genomic DNA. MtDNA encodes two forms of mitochondrial ribosomal RNA (12 S RNA and 16 S RNA) and 22 tRNAs as well as 13 polypeptides that serve as subunits to form the mitochondrial respiratory chain complex. Human mitoribosomes have a sedimentation coefficient of 55 S, consisting of a 39 S large subunit and a 28 S small subunit. Thirteen proteins in the human mitoribosome subunits are homologous to those in prokaryotes, while the remaining eight are unique to mammals (Suzuki et al., 2001). Based on proteomics analyses of mitoribosomes in yeast and mammals, DAP3 has been identified as a component of the mitoribosome 28 S subunit (Cavdar Koc et al., 2001; Suzuki et al., 2001; Saveanu et al., 2001). An important feature of the mitoribosome 28 S small subunit is the involvement of its inherent GTP enzyme activity in initiating mitoribosome translation. DAP3 is the only protein with GTP enzyme activity reported in the 28 S subunit and cooperates with other ribosome subunits to translate mitochondrial-encoded proteins (Cavdar Koc et al., 2001; O’Brien, 2003). The deletion of DAP3 inhibited the synthesis of mtDNA-encoded proteins, such as COX I and COX II. Additionally, deletion of the respiratory chain complex subunit reduced the oxidative phosphorylation capacity of mitochondria and diminished the capacity for ATP synthesis (Xiao et al., 2015; Jacques et al., 2006; Kim et al., 2007). Evidence suggests that DAP3 plays a role in mitochondrial protein and regulation of cellular energy metabolism.
DAP3 and cellular redox balance
Mitochondria are the main sites of ATP production, with reactive oxygen species (ROS) formed as by-products of redox reactions (Guo et al., 2024; Musicco et al., 2023). Under physiological conditions, intracellular ROS are rapidly eliminated by an intracellular reduction system comprising reduced nicotinamide adenine dinucleotide phosphate (NADPH) and superoxide dismutase (SOD) (Boonstra & Post, 2004; Malik & Herbert, 2012). However, when ROS production exceeds the scavenging capacity of the intracellular reduction system, it can lead to oxidative damage to cells and organelles, resulting in oxidative stress-related diseases, such as diffuse large B-cell lymphoma (Yin et al., 2023; Feng et al., 2023). Studies have shown that hydrogen peroxide (H2O2) significantly reduces the expression of cellular DAP3, while radiation has no effect on its expression (Murata et al., 2006). ROS production was decreased after downregulation of DAP3 expression in NIH 3T3 cells (Murata et al., 2006). Furthermore, the sensitivity of the cell to H2O2 is reduced following downregulation of DAP3 expression, while exogenous DAP3 overexpression has the reverse effect (Boonstra & Post, 2004; Malik & Herbert, 2012; Murata et al., 2006). Thus, evidence suggests that downregulation of DAP3 enhances cellular resistance to oxidative stress and reduces excessive ROS production, thereby contributing to the regulation of cellular redox balance.
DAP3 regulates apoptosis
Initial research demonstrated that treatment of HeLa cells with interferon-γ (IFNγ) induced significant apoptosis, and that this IFNγ sensitivity reduced following downregulation of DAP3. However, in IFNγ untreated cells, no apoptosis was observed, even when DAP3 expression was downregulated (Kissil et al., 1995; Kissil & Kimchi, 1998), indicating that DAP3 is crucial for IFNγ-induced apoptosis. Furthermore, exogenous overexpression of DAP3 effectively induced programmed cell death (PCD) (Kissil et al., 1995; Kissil & Kimchi, 1998). Therefore, DAP3 is regarded as an important member of the death-related protein family.
The mechanism of apoptosis in eukaryotic cells is divided into exogenous and endogenous pathways (Wazir et al., 2015). The exogenous apoptosis pathway is triggered mainly by the combination of TNFα, factor-associated suicide (Fas), TNF-related apoptosis-inducing ligand (TRAIL), and other receptors and ligands of the TNF family. These combinations lead to the recruitment of caspase-8 and caspase-10 precursors to form a death-inducing signaling complex (DISC), which activates cell death proteases to induce apoptosis (Wazir et al., 2015). As an adapter protein, DAP3 participates directly in the exogenous cell apoptosis pathway mediated by TNF-α and Fas-L (Fig. 2). In addition, DAP3 binds to the death signal enhancer (DELE), thereby mediating the interaction of TRAIL with FADD and participating in the exogenous cell apoptosis pathway. In HeLa cells, DELE was shown to bind to DAP3, connecting the TRAIL receptor with FADD to activate the DISC, which includes the caspase-8 precursor. Downregulation of DELE expression inhibited apoptosis induced by TNF-α, Fas, and TRAIL. Moreover, specific downregulation of DAP3 expression via RNA interference technology also inhibited Fas-mediated apoptosis, while this process was promoted by exogenous overexpression of DAP3 (Kissil et al., 1999; Miyazaki & Reed, 2001; Harada et al., 2010).
Fig. 2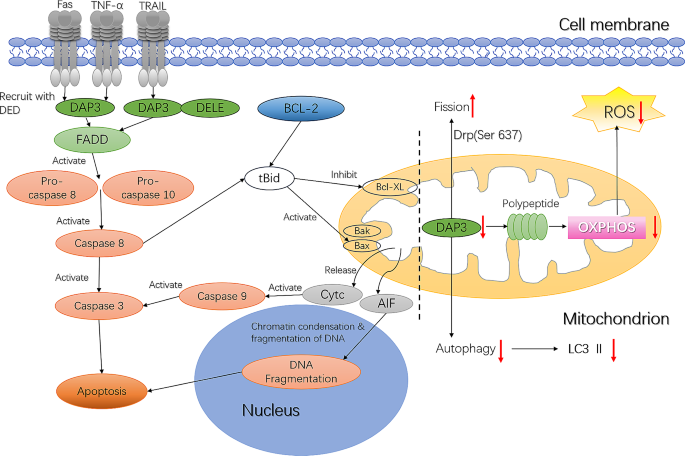
The function of DAP3 within the mitochondria and the DAP3-mediated apoptosis pathway
In the endogenous apoptosis pathway, mitochondrial stimulators (such as stress conditions, chemotherapy drugs, etc.) recruit pro-apoptotic protein from the B-cell lymphoma 2 (Bcl2) family to the mitochondrial outer membrane, leading to an increase in mitochondrial outer membrane permeability and promoting the release of pro-apoptotic proteins such as cytochrome c (Cytc) and apoptosis-inducing factor (AIF) into the cytoplasm from the mitochondrial membrane gap. Once released into the cytoplasm, Cytc initiates apoptosis by binding to apoptotic protease activating factor-1 (APAF-1) (caspase-9 activator) (Fox et al., 2016; Kiraz et al., 2016), while AIF is transported into the nucleus where it accumulates and activates endogenous endonucleases, which cleave DNA into 50-kb fragments, ultimately triggering apoptosis (Meggyeshazi et al., 2014). The opening of the permeability transition pore in mitochondria during apoptosis results in dissipation of the inner mitochondrial membrane potential, production of reactive oxygen species (ROS), and reduction in ATP synthesis. DAP3 located within mitochondria mediates mitochondrial fragmentation, leading to a decrease in membrane potential, opening of permeability transition pores, and release of mitochondrial components such as Cytc into the cytoplasm. However, DAP3 was the first mitochondrial matrix protein shown to be involved in the regulation of cell apoptosis, but not released from the mitochondria. Thus, the correlation between DAP3 and endogenous cell apoptosis and the specific mechanism underlying its role in this process remain to be clarified (Mukamel & Kimchi, 2004).
DAP3 promotes apoptosis mediated via the T cell receptor (TCR) in immature thymocytes to achieve negative selection of autoreactive cells during thymic development (Tosa et al., 2010). In this process, orphan nuclear receptor 77 (Nur77), which is a member of the steroid/thyroid hormone receptor superfamily, is activated via the TCR and the transcript is subsequently transported to the nucleus by binding to the N-terminal domain of DAP3, thereby participating in TCR-mediated cell apoptosis (Tosa et al., 2010). DAP3 is expressed in CD4+, CD8+ double-positive T cells as well as in CD4+, CD8+ single-positive T cells, but not in CD4−, CD8− double-negative T cells (Tosa et al., 2010). The presence of CD4+, CD8+ double-positive T cells during thymic development triggers apoptosis of these cells to prevent self-attack (Tosa et al., 2010). DAP3-mediated cell negative selection still occurred following mutation of the DAP3 domain (the region that binds to FADD), indicating that DAP3 may not function through FADD in T cells (Tosa et al., 2010).
DAP3 and anoikis
Anoikis is a programmed cell death process that occurs when cells lose their attachment to the extracellular matrix (ECM) or neighboring cells. This phenomenon is crucial for maintaining tissue homeostasis, as it acts as a protective mechanism against the survival and dissemination of detached or misplaced cells (Lee et al., 2021). Anoikis is induced by the activation of caspase-8 mediated by the joint action of DAP3 and FADD (Li et al., 2009). Co-immunoprecipitation studies showed that the DAP3-FADD interaction is minimal when the cells adhere to the ECM, and this interaction increases significantly when the cells are in suspension (Miyazaki et al., 2004). The interaction between DAP3 and FADD was found to be significantly reduced by administration of exogenous protein kinase B (AKT) activators (Li et al., 2009). The interaction between FADD and DAP3 also involves interferon-β promoter stimulator 1 (IPS-1) (Li et al., 2009), which is an integral component of the innate immune system. Downregulated IPS-1 expression inhibited DAP3-mediated anoikis, but did not affect IPS-1 mediated anoikis (Li et al., 2009). In addition, reduced endogenous DAP3 expression inhibited cell anoikis, while exogenous overexpression of DAP3 promoted cell detachment from the survival matrix and accelerated cell death. AKT has also been shown to play a crucial role in inhibiting DAP3-induced cell anoikis and protecting anchorage-independent survival of transformed cells (Franke et al., 2003; Miyazaki et al., 2004). Indeed, DAP3-induced cell anoikis is attributed primarily to AKT or closely related enzymes that promote phosphorylation of DAP3 threonine at position 273, which hinders the interaction between DAP3 and FADD, ultimately obstructing the exogenous apoptosis pathway (Miyazaki et al., 2004). DAP3 contains the RXRXX (S/T) structure necessary for AKT-mediated substrate phosphorylation. Mutation of this structural motif led to the loss of DAP3 phosphorylation and increased cell death caused by anoikis (Miyazaki et al., 2004). Thus, the role of DAP3 in anoikis represents an additional mechanism underlying its participation in the exogenous apoptosis pathway.
DAP3 and autophagy
Autophagy refers to the process of self-degradation and reuse of intracellular components under conditions of starvation, stress, and growth factor deficiency that occurs widely in eukaryotic cells (Klionsky et al., 2021; Debnath et al., 2023). Autophagy begins with the formation of an autophagosome, which encapsulates cell components, including long-lived proteins, damaged organelles, and pathogens, from the cell membrane structure. Subsequently, the outer membrane of the autophagosome combines with the lysosomal membrane, lysosome hydrolases degrade its contents, and the decomposed products are reused by the cell (Miller & Thorburn, 2021). Phagophore extension, which is essential for maturation of the autophagosome, relies on two sumoylation systems. Each system involves microtubules association protein 1 light chain 3 (LC3/ATG8), autophagy-related protein 12 (ATG12) and three enzymes necessary for ligation reaction: ATG3, ATG7, and ATG10. LC3 is the main index used to monitor autophagy, which plays a significant role in various physiological and pathological processes, including tumors, neurodegeneration, metabolic diseases, and cardiovascular diseases (Marsh & Debnath, 2020; Festa et al., 2023; Wang et al., 2023; Popov et al., 2023). Members of the death-related protein family, such as DAP1 (Koren et al., 2010), participate in autophagy regulation. Recent studies have shown that mitochondria are the key to autophagy regulation (Graef & Nunnari, 2011; Hailey et al., 2010). Lin et al. conducted immunofluorescence studies that showed an increase in LC3 fluorescent spots in serum-starved cells compared with cells without serum starvation, and that this effect was lost in cells with downregulated DAP3 expression (Xiao et al., 2015). Furthermore, Western blot analysis demonstrated that when DAP3 protein expression decreased, the level of LC3II decreased regardless of whether the cells were exposed to starvation conditions or not. These findings indicate that DAP3 expression is required for autophagy.
DAP3 and tumors
Studies have indicated the role of apoptosis in tumor occurrence and development, which has led to more focused research on the underlying mechanism (Zhang et al., 2022; Zhao et al., 2023). Various anticancer drugs exert their effects by inducing tumor cell apoptosis (Lu et al., 2022; Chimento et al., 2023). For example, curcumol induces breast cancer cell apoptosis by downregulating the expression of mutant P53, and also induces colorectal cancer cell apoptosis by increasing P38 phosphorylation (Wang et al., 2015). Metformin combined with oxaliplatin induces apoptosis of the gastric cancer cell lines SGC7901 and SNU-16 by upregulating expression of the apoptosis-related proteins Bax and caspase-3 and downregulating expression of the anti-apoptotic protein Bcl-2 (Zhu et al., 2022). The combination of ivermectin and gemcitabine induces excessive ROS production and mitochondrial dysfunction, while reducing the mitochondrial membrane potential, and promoting pancreatic cancer cell apoptosis (Lee et al., 2022).
To date, both tumor-promoting and tumor-suppressing effects of DAP3 have been reported (Han et al., 2022). Compared with normal tissues, DAP3 transcript levels have been detected at significantly higher levels in pancreatic cancer tissues. Furthermore, patients with high levels of DAP3 exhibited a significantly shorter overall survival compared to patients with low levels (Sui et al., 2021). It has also been observed that DAP3 protein and mRNA levels are increased in invasive malignant glioma and glioma cell lines with induced motility phenotype (Mariani et al., 2001). Upregulated levels of DAP3 protein decreased camptothecin-induced apoptosis, while downregulated DAP3 levels reduced cell migration (Mariani et al., 2001). These findings indicate that DAP3 inhibits tumor cell apoptosis and maintains the migration ability of invasive malignant glioma cells.
The nuclear transcription factor Ikaros plays a significant role in the development and differentiation of normal lymphocytes (Schwickert et al., 2019). Studies have shown a high frequency of Ikaros inactivation in human and mouse lymphomas, which may contribute to lymphoma development (Yasumura et al., 2004). DAP3 expression is upregulated in mouse 3T3-L1 cells with Ikaros overexpression (Yasumura et al., 2004). In lymphoma cells, DAP3 gene expression is decreased due to the absence of the Ikaros gene, which allows tumor cells to resist the exogenous apoptotic signaling pathway mediated by TNF-α, ultimately leading to their survival (Yasumura et al., 2004).
Liver kinase B1 (LKB1), a tumor suppressor involved in regulation of the apoptotic pathway, binds to LKB1 interacting protein 1 (LIP1) in an interaction that anchors it in the cytoplasm (Wazir et al., 2015). In osteosarcoma cells, LKB1 interacts with DAP3 through LIP1. Co-expression of LKB1 and DAP3 was shown to significantly reduce osteosarcoma cell colony formation and induce apoptosis, while the loss of LKB1 activity reduced the apoptosis induced by DAP3 (Takeda et al., 2007; Smith et al., 2001). However, co-overexpression of LKB1 and a DAP3 mutant (dominant inactivated structural mutation) increased osteosarcoma cell colony formation (Takeda et al., 2007). Thus, it is speculated that LKB1 and DAP3 jointly regulate osteosarcoma cell death via the exogenous apoptosis pathway (Takeda et al., 2007).
DAP3 mRNA levels in stage IV thymoma patients were reported to be significantly higher than those in stage I patients, suggesting that DAP3 may be involved in the occurrence and development of thymoma (Sasaki et al., 2004). Indeed, DAP3 protein expression levels are related to the clinical classification of thymoma by the World Health Organization (WHO). In thyroid eosinophilic adenoma, both the protein and mRNA levels of DAP3 were higher than those in the control group. Moreover, high DAP3 expression was associated with a low rate of apoptosis (Wazir et al., 2012). In a study of DAP3 mRNA levels in 153 breast cancer patients that included a 10-year follow-up, researchers found that patients without in situ recurrence had relatively higher DAP3 transcript levels compared to patients with in situ recurrence or distant metastasis (Wazir et al., 2012). In addition, DAP3 transcription levels correlated with TNM (T: topography, N: lymph node, M: Metastasis) staging of breast cancer (Wazir et al., 2012), with lower levels in TNM stage 3 patients compared to those with stage 1 disease (Wazir et al., 2012). Kaplan–Meier survival analysis has shown that patients with higher DAP3 mRNA levels have a better prognosis, possibly due to the pro-apoptotic effect of DAP3. Conversely, low DAP3 expression is associated with local recurrence, distant metastasis, and mortality (Wazir et al., 2012). In gastric cancer, DAP3 protein and mRNA levels are lower than those in normal tissues. DAP3 downregulation may be linked to distant metastasis and can contribute to chemotherapy drug resistance in patients (Jia et al., 2014). Similarly, downregulated DAP3 expression modified the cellular radiation response of lung adenocarcinoma cells through the retinoic acid-inducible gene-I (RIG-I)-like receptor (RLR) agonist Poly(I:C) (Sato et al., 2021). DAP3 also plays an important role in the regulation of G2/M arrest via pChk1 in irradiated human lung adenocarcinoma cells suggesting that the Chk1-mediated G2/M arrest is a key regulator of radioresistance in H1299 and A549cells (Sato et al., 2023).
Sui et al. reported that the levels of DAP3 and DELE1 exhibited a strong correlation with the clinical treatment effect and recurrence in human colorectal cancer, with levels of DAP3 at the transcription level, and high expression of both DAP3 and DELE1 at the protein level (Sui et al., 2023). DAP3 and DELE1 have been identified as potential prognostic indicators for colorectal cancer. Additionally, in vitro experiments have shown that knockdown of DAP3 and DELE1 alone or in combination improve sensitivity to chemotherapy (5‑FU, docetaxel, methotrexate). According to RNA-seq data from The Cancer Genome Atlas database, DAP3 is highly expressed in 17 tumor types (Han et al., 2020). DAP3 knockdown in ESCC and HCC cells inhibited the tumor cell phenotype, while the phenotype was restored following DAP3 re-expression, indicating that DAP3 functions as an oncogene. A-to-I RNA editing, a post-transcriptional modification in which adenosines are converted to inosines in both coding and noncoding RNA transcript by double-stranded RNA-specific adenosine deaminase (ADAR) enzymes. In normal cells, this process requires recognition and binding of dsDNA by the ADAR2 deaminase domain. However, in cancer cells, DAP3 interacts preferentially with the ADAR2 deaminase domain, which inhibits the combination of ADAR2 and dsRNA subunit, thereby promoting tumor development (Han et al., 2020).
Variations in the expression of DAP3 have been reported in different types of tumors (Table 1), indicating differences in the balance between the role of DAP3 in maintaining mitochondrial energy metabolism and promoting apoptosis across tumor types. Further analysis is required to determine which effect dominates based on the characteristics of each specific tumor type.
Tumor DAP3 mRNA DAP3 protein Reference | ||
|---|---|---|
Glioblastoma (Mariani et al., 2001) | ↑ | ↑ |
Thymoma (Sasaki et al., 2004) | ↑ | |
Thyroid oncocytic tumours (Jacques et al., 2009) | ↑ | ↑ |
Breast cancer (Wazir et al., 2012) | ↓ | |
Gastric cancer | ↓ | ↓ |
Pancreatic Cancer (Sui et al., 2021) | ↑ |
Table 1 DAP3 expression in different tumors
Perspectives
As a component of the mitochondrial ribosome 28 S subunit, DAP3 plays a vital role in the exogenous apoptosis pathway through its interaction with glucocorticoid receptors to regulate mitochondrial redox balance, mitochondrial dynamics, and autophagy. However, DAP3 plays different roles in various tumors, which may depend on the genetic background, microenvironment, and heterogeneity of tumors. The pro-apoptotic function of DAP3 makes it a potential target for anti-tumor; however, its role in synthesizing respiratory chain proteins to provide energy for tumor cells necessitates a re-evaluation of its significance in tumor treatment. Therefore, future research should extensively analyze the expression and function of DAP3 in different tumors and explore its involvement in cellular signaling pathways, with the aim of establishing DAP3 as a potential diagnostic, prognostic and therapeutic target for specific types of tumor.
Data availability
Not applicable.
References
Berger, T., & Kretzler, M. (2002). Interaction of DAP3 and FADD only after cellular disruption. Nature Immunology, 3(1), 3–5. https://doi.org/10.1038/ni0102-3b.
Boonstra, J., & Post, J., A (2004). Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene, 337, 1–13. https://doi.org/10.1016/j.gene.2004.04.032.
Cavdar, K. E., Ranasinghe, A., Burkhart, W., Blackburn, K., Koc, H., Moseley, A., & Spremulli, L. L. (2001). A new face on apoptosis: Death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Letters, 492(1–2), 166–170. https://doi.org/10.1016/s0014-5793(01)02250-5.
Chimento, A., Arianna, D. L., D’Amico, M., Francesca, D., A., & Pezzi, V. (2023). The involvement of natural polyphenols in molecular mechanisms inducing apoptosis in tumor cells: A promising adjuvant in Cancer Therapy. International Journal of Molecular Sciences, 24(2), 1680. https://doi.org/10.3390/ijms24021680.
Debnath, J., Gammoh, N., & Ryan, K. M. (2023). Autophagy and autophagy-related pathways in cancer. Nature Reviews Molecular Cell Biology, 24(8), 560–575. https://doi.org/10.1038/s41580-023-00585-z.
Feng, Q. L., Hu, K., Hu, H., Lu, Y., Zhang, H., Wang, G. L., Zhang, Q., Xu, Z., Gao, X. J., Jia, X., Zhu, H., Song, D., Yi, H., Peng, Y., Wu, X., Li, B., Zhu, W., & Shi, J. (2023). Berberine derivative DCZ0358 induce oxidative damage by ROS-mediated JNK signaling in DLBCL cells. International Immunopharmacology, 125(Pt A). https://doi.org/10.1016/j.intimp.2023.111139.
Festa, B. P., Siddiqi, F. H., Jimenez-Sanchez, M., Won, H., Rob, M., Djajadikerta, A., Stamatakou, E., & Rubinsztein, D., C (2023). Microglial-to-neuronal CCR5 signaling regulates autophagy in neurodegeneration. Neuron, 111(13), 2021–2037. https://doi.org/10.1016/j.neuron.2023.04.006.
Fox, L., Joanna, M. F., & Marion (2016). Targeting cell death signalling in cancer: Minimising ‘Collateral damage’. British Journal of Cancer, 115(1), 5–11. https://doi.org/10.1038/bjc.2016.111.
Franke, T., Hornik, C., Segev, L., Shostak, G., & Sugimoto, C. (2003). PI3K/Akt and apoptosis: Size matters. Oncogene, 22(56), 8983–8998. https://doi.org/10.1038/sj.onc.1207115.
Graef, M., & Nunnari, J. (2011). Mitochondria regulate autophagy by conserved signalling pathways. EMBO Journal, 30(11), 2101–2114. https://doi.org/10.1038/emboj.2011.104.
Guo, J., Yang, N., Zhang, J., Huang, Y., Xiang, Q., Wen, J., Chen, Y., Hu, T., Qiuyan, L., & Rao, C. (2024). Neurotoxicity study of ethyl acetate extract of Zanthoxylum armatum DC. On SH-SY5Y based on ROS mediated mitochondrial apoptosis pathway. Journal of Ethnopharmacology, 319(Pt3), 117321. https://doi.org/10.1016/j.jep.2023.117321.
Hailey, D. W., Rambold, A. S., Satpute-Krishnan, P., Mitra, K., Sougrat, R., Kim, P., K., & Lippincott-Schwartz, J. (2010). Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell, 141(4), 656–667. https://doi.org/10.1016/j.cell.2010.04.009.
Han, J., An, O., Hong, H., Chan, T. H., M, & Chen, L. (2020). Suppression of adenosine-to-inosine (A-to-I) RNA editome by death associated protein 3 (DAP3) promotes cancer progression. Science Advances, 6(25), eaba5136. https://doi.org/10.1126/sciadvaba5136.
Han, J., An, O., Ren, X., Song, Y., Tang, S. J., Shen, H., Ke, X., Ng, V. H. E., Tay, D. J. T., Tan, H. Q., Kappei, D., Yang, H., & Chen, L. (2022). Multilayered control of splicing regulatory networks by DAP3 leads to widespread alternative splicing changes in cancer. Nature Communications, 13(1), 1793. https://doi.org/10.1038/s41467-022-29400-7.
Harada, T., Iwai, A., & Miyazaki, T. (2010). Identification of DELE, a novel DAP3-binding protein which is crucial for death receptor-mediated apoptosis induction. Apoptosis, 15(10), 1247–1255. https://doi.org/10.1007/s10495-010-0519-3.
Hulkko, S. M., & Zilliacus, J. (2002). Functional interaction between the pro-apoptotic DAP3 and the glucocorticoid receptor. Biochemical and Biophysical Research Communications, 295(3), 749–755. https://doi.org/10.1016/S0006-291X(02)00713-1.
Hulkko, S. M., Wakui, H., & Zilliacus, J. (2000). The pro-apoptotic protein death-associated protein 3 (DAP3) interacts with the glucocorticoid receptor and affects the receptor function. Biochemical Journal, 349(Pt 3), 885–893. https://doi.org/10.1042/bj3490885.
Jacques, C., Chevrollier, A., Loiseau, D., Lagoutte, L., Savagner, F., Malthiery, Y., & Reynier, P. (2006). mtDNA controls expression of the Death Associated protein 3. Experimental Cell Research, 312(6), 737–745. https://doi.org/10.1016/j.yexcr.2005.11.027.
Jacques, C., Fontaine, J., Franc, F., Mirebeau-Prunier, B., Triau, D., Savagner, S., F., & Malthiery, F. (2009). Death-associated protein 3 is overexpressed in human thyroid oncocytic tumours. British Journal of Cancer, 101(1), 132–138. https://doi.org/10.1038/sj.bjc.6605111.
Jia, Y., Ye, L., Ji, K., Zhang, L., Hargest, R., Ji, J., & Jiang, W., J (2014). Death-associated protein-3, DAP-3, correlates with preoperative chemotherapy effectiveness and prognosis of gastric cancer patients following perioperative chemotherapy and radical gastrectomy. British Journal of Cancer, 110(2), 421–429. https://doi.org/10.1038/bjc.2013.712.
Kim, H. R., Chae, H. J., Thomas, M., Miyazaki, T., Monosov, A., Monosov, E., Krajewska, M., Krajewski, S., & Reed, J. C. (2007). Mammalian dap3 is an essential gene required for mitochondrial homeostasis in vivo and contributing to the extrinsic pathway for apoptosis. The FASEB Journal, 21(1), 188–196. https://doi.org/10.1096/fj.06-6283com.
Kiraz, Y., Adan, A., Kartal, Y. M., & Baran, Y. (2016). Major apoptotic mechanisms and genes involved in apoptosis. Tumour Biology, 37(7), 8471–8486. https://doi.org/10.1007/s13277-016-5035-9.
Kissil, J. L., & Kimchi, A. (1997). Assignment of death associated protein 3 (DAP3) to human chromosome 1q21 by in situ hybridization. Cytogenetics and Cell Genetics, 77(3–4). https://doi.org/10.1159/000134587.
Kissil, J. L., & Kimchi, A. (1998). Death-associated proteins: From gene identification to the analysis of their apoptotic and tumour suppressive functions. Molecular Medicine Today, 4(6), 268–274. https://doi.org/10.1016/S1357-4310(98)01263-5.
Kissil, J. L., Deiss, L. P., Bayewitch, M., Raveh, T., Khaspekov, G., & Kimchi, A. (1995). Isolation of DAP3, a novel mediator of interferon-gamma-induced cell death. Journal of Biological Chemistry, 270(46), 27932–27936. https://doi.org/10.1074/jbc.270.46.27932.
Kissil, J. L., Cohen, O., Raveh, T., & Kimchi, A. (1999). Structure-function analysis of an evolutionary conserved protein, DAP3, which mediates TNF-alpha - and Fas-induced cell death. EMBO Journal, 18(2), 353–362. https://doi.org/10.1093/emboj/18.2.353.
Klionsky, D. J., Petroni, G., Amaravadi, R. K., Baehrecke, E. H., Ballabio, A., Boya, P., Bravo-San, P. J., Cadwell, M., Cecconi, K., Choi, F., Choi, A. M. K., Chu, M. E., Codogno, C. T., Colombo, P., Cuervo, M. I., Deretic, A. M., Dikic, V., Elazar, I., Eskelinen, Z., … Pietrocola, F. (2021). Autophagy in major human diseases. EMBO Journal, 40(19). https://doi.org/10.15252/embj.2021108863.
Koren, I., Reem, E., & Kimchi, A. (2010). DAP1, a novel substrate of mTOR, negatively regulates autophagy. Current Biology, 20(12), 1093–1098. https://doi.org/10.1016/j.cub.2010.04.041.
Lee, H. Y., Son, S. W., Moeng, S., Choi, S. Y., & Park, J. K. (2021). The role of noncoding RNAs in the regulation of anoikis and anchorage-independent growth in cancer. International Journal of Molecular Sciences, 22(2). https://doi.org/10.3390/ijms22020627.
Lee, D., Kang, H. W., Kim, S. Y., Kim, M. J., Jeong, J. W., Hong, W. C., Fang, S., Kim, H. S., Lee, Y. S., Kim, H. J., & Park, J. S. (2022). Ivermectin and gemcitabine combination treatment induces apoptosis of pancreatic cancer cells via mitochondrial dysfunction. Frontiers in Pharmacology, 13, 934746. https://doi.org/10.3389/fphar.2022.934746.
Lee, H., Lee, T. J., Galloway, C. A., Zhi, W., Xiao, W., Mesy, B. K. L., Sharma, A., Teng, Y., Sesaki, H., & Yoon, Y. (2023). The mitochondrial fusion protein OPA1 is dispensable in the liver and its absence induces mitohormesis to protect liver from drug-induced injury. Nature Communications, 14(1), 6721–6721. https://doi.org/10.1038/S41467-023-42564-0.
Li, H. M., Fujikura, D., Harada, T., Uehara, J., Kawai, K., Akira, S., Reed, J. C., Iwai, A., & Miyazaki, T. (2009). IPS-1 is crucial for DAP3-mediated anoikis induction by caspase-8 activation. Cell Death & Differentiation, 16(2), 1615–1621. https://doi.org/10.1086/520246.
Lu, J., Li, Y., Gong, S., Wang, J., Lu, X., Jin, Q., Lu, B., & Chen, Q. (2022). Ciclopirox targets cellular bioenergetics and activates ER stress to induce apoptosis in non-small cell lung cancer cells. Cell Communication and Signaling, 20(1), 37. https://doi.org/10.1186/s12964-022-00847-x.
Malik, Q., & Herbert, K. E. (2012). Oxidative and non-oxidative DNA damage and cardiovascular disease. Free Radical Research, 46(4), 554–564. https://doi.org/10.3109/10715762.2012.663913.
Mariani, L., Beaudry, C., Mcdonough, W., Hoelzinger, S., Kaczmarek, D. B., Ponce, E., Coons, F., Giese, S. W., Seiler, A., R. W., & Berens, M. E. (2001). Death-associated protein 3 (Dap-3) is overexpressed in invasive glioblastoma cells in vivo and in glioma cell lines with induced motility phenotype in vitro. Clinical Cancer Research, 7(8), 2480–2489. https://doi.org/10.1093/carcin/22.8.1327.
Marsh, T., & Debnath, J. (2020). Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy, 16(6), 1164–1165. https://doi.org/10.1080/15548627.2020.1753001.
Meggyeshazi, N., Andocs, G., Balogh, L., Balla, P., Kiszner, G., Teleki, I., Jeney, A., & Krenacs, T. (2014). DNA fragmentation and caspase-independent programmed cell death by modulated electrohyperthermia. Strahlentherapie Und Onkologie, 190(9), 815–822. https://doi.org/10.1007/s00066-014-0617-1.
Miller, D. R., & Thorburn, A. (2021). Autophagy and organelle homeostasis in cancer. Developmental Cell, 56(7), 906–918. https://doi.org/10.1016/j.devcel.2021.02.010.
Miller, J. L., Koc, H., & Koc, E., C (2008). Identification of phosphorylation sites in mammalian mitochondrial ribosomal protein DAP3. Protein Science, 17(2), 251–260. https://doi.org/10.1110/ps.073185608.
Miyazaki, T., & Reed, J. C. (2001). A GTP-binding adapter protein couples TRAIL receptors to apoptosis-inducing proteins. Nature Immunology, 2(6), 493–500. https://doi.org/10.1038/88684.
Miyazaki, T., Shen, M., Fujikura, D., Tosa, N., Kim, H. R., Kon, S., Uede, T., & Reed, J. C. (2004). Functional role of death-associated protein 3 (DAP3) in anoikis. Journal of Biological Chemistry, 279(43), 44667–44672. https://doi.org/10.1074/jbc.M408101200.
Morgan, C. J., Jacques, C., Savagner, F., Tourmen, Y., Mirebeau, D. P., Malthiery, Y., & Reynier, P. (2001). A conserved N-terminal sequence targets human DAP3 to mitochondria. Biochemical and Biophysical Research Communications, 280(1), 177–181. https://doi.org/10.1006/bbrc.2000.4119.
Mukamel, Z., & Kimchi, A. (2004). Death-associated protein 3 localizes to the mitochondria and is involved in the process of mitochondrial fragmentation during cell death. Journal of Biological Chemistry, 279(35), 36732–36738. https://doi.org/10.1074/jbc.M400041200.
Murata, Y., Wakoh, T., Uekawa, N., Sugimoto, M., Asai, A., Miyazaki, T., & Maruyama, M. (2006). Death-associated protein 3 regulates cellular senescence through oxidative stress response. FEBS Letters, 580(26), 6093–6099. https://doi.org/10.1016/j.febslet.2006.10.004.
Musicco, C., Signorile, A., Pesce, V., Loguercio, P. P., & Cormio, A. (2023). Mitochondria deregulations in cancer offer several potential targets of therapeutic interventions. International Journal of Molecular Sciences, 24(13), 10420. https://doi.org/10.3390/IJMS241310420.
O’Brien, T. W. (2003). Properties of human mitochondrial ribosomes. IUBMB Life, 55(9), 505–513. https://doi.org/10.1080/15216540310001626610.
Popov, S. V., Mukhomedzyanov, A. V., Voronkov, N. S., Derkachev, I. A., Boshchenko, A. A., Fu, F., Sufianova, G., Khlestkina, Z., M,S., & Maslov, L., N (2023). Regulation of autophagy of the heart in ischemia and reperfusion. Apoptosis, 28(1–2), 55–80. https://doi.org/10.1007/s10495-022-01786-1.
Sasaki, H., Ide, N., Yukiue, H., Kobayashi, Y., Fukai, I., Yamakawa, Y., & Fujii, Y. (2004). Arg and DAP3 expression was correlated with human thymoma stage. Clinical & Experimental Metastasis, 21(6), 507–513. https://doi.org/10.1007/s10585-004-2153-3.
Sato, Y., Yoshino, H., Kashiwakura, I., & Tsuruga, E. (2021). DAP3 is involved in modulation of cellular radiation response by RIG-I-Like receptor agonist in human lung adenocarcinoma cells. International Journal of Molecular Sciences, 22(1), 420. https://doi.org/10.3390/ijms22010420.
Sato, Y., Yoshino, H., Sato, K., Kashiwakura, I., & Tsuruga, E. (2023). DAP3-mediated cell cycle regulation and its association with radioresistance in human lung adenocarcinoma cell lines. Journal of Radiation Research, 64(3), 520–529. https://doi.org/10.1093/jrr/rrad016.
Saveanu, C., Fromont-Racine, M., Harington, A., Ricard, F., Namane, A., & Jacquier, A. (2001). Identification of 12 new yeast mitochondrial ribosomal proteins including 6 that have no prokaryotic homologues. Journal of Biological Chemistry, 276(19), 15861–15867. https://doi.org/10.1074/jbc.M010864200.
Schwickert, T. A., Tagoh, H., Schindler, K., Fischer, M., Jaritz, M., & Busslinger, M. (2019). Ikaros prevents autoimmunity by controlling anergy and toll-like receptor signaling in B cells. Nature Immunology, 20(11), 1517–1529. https://doi.org/10.1038/s41590-019-0490-2.
Smith, D. P., Rayter, S., Niederlander, I., Spicer, G., Jones, J.,C, M., & Ashworth, A. (2001). LIP1, a cytoplasmic protein functionally linked to the Peutz-Jeghers syndrome kinase LKB1. Human Molecular Genetics, 10(25), 2869–2877. https://doi.org/10.1093/hmg/10.25.2869.
Sui, L., Ye, L., Sanders, A., Yang, J., Hao, Y., Hargest, C. R., & Jiang, W. G. (2021). Expression of death associated proteins DAP1 and DAP3 in human pancreatic cancer. Anticancer Research, 41(5), 2357–2362. https://doi.org/10.21873/anticanres.15010.
Sui, L., Zeng, J., Zhao, H., Ye, L., Martin, T. A., Sanders, A., Ruge, J., Jiang, F., Dou, A., Hargest, Q. P., Song, R. X., & Jiang, W., J (2023). Death associated protein–3 (DAP3) and DAP3 binding cell death enhancer–1 (DELE1) in human colorectal cancer, and their impacts on clinical outcome and chemoresistance. International Journal of Oncology, 62(1), 7. https://doi.org/10.3892/ijo.2022.5455.
Suzuki, T., Terasaki, M., Takemoto-Hori, C., Hanada, T., Ueda, T., Wada, A., & Watanabe, K. (2001). Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. Journal of Biological Chemistry, 276(35), 33181–33195. https://doi.org/10.1074/jbc.M103236200.
Takeda, S., Iwai, A., Nakashima, M., Fujikura, D., Chiba, S., Li, H. M., Uehara, J., Kawaguchi, S., Kaya, M., Nagoya, S., Wada, T., Yuan, J., Rayter, S., Ashworth, A., Reed, J., Yamashita, C., Uede, T., T., & Miyazaki, T. (2007). LKB1 is crucial for TRAIL-mediated apoptosis induction in osteosarcoma. Anticancer Research, 27(2), 761–768. https://doi.org/10.1021/cm059988s.
Tosa, N., Iwai, A., Tanaka, T., Kumagai, T., Nitta, T., Chiba, S., Maeda, M., Takahama, Y., Uede, T., & Miyazaki, T. (2010). Critical function of death-associated protein 3 in T cell receptor-mediated apoptosis induction. Biochemical and Biophysical Research Communications, 395(3), 356–360. https://doi.org/10.1016/j.bbrc.2010.04.018.
Wang, J., Huang, F., Bai, Z., Chi, B., Wu, J., & Chen, X. (2015). Curcumol inhibits growth and induces apoptosis of colorectal cancer LoVo Cell line via IGF-1R and p38 MAPK pathway. International Journal of Molecular Sciences, 16(8), 19851–19867. https://doi.org/10.3390/ijms160819851.
Wang, J., Zhang, Y., Cao, J., Wang, Y., Anwar, N., Zhang, Z., Zhang, D., Ma, Y., Xiao, Y., Xiao, L., & Wang, X. (2023). The role of autophagy in bone metabolism and clinical significance. Autophagy, 19(9), 2409–2427. https://doi.org/10.1080/15548627.2023.2186112.
Wazir, U., Jiang, W. G., Sharma, A. K., & Mokbel, K. (2012). The mRNA expression of DAP3 in human breast cancer: Correlation with clinicopathological parameters. Anticancer Research, 32(2), 671–674. https://doi.org/10.1007/s10014-011-0075-8.
Wazir, U., Orakzai, M. M., Khanzada, Z. S., Jiang, W. G., Sharma, A. K., Kasem, A., & Mokbel, K. (2015). The role of death-associated protein 3 in apoptosis, anoikis and human cancer. Cancer Cell International, 15, 39. https://doi.org/10.1186/s12935-015-0187-z.
Xiao, L., Xian, H., Lee, K. Y., Xiao, B., Wang, H., Yu, F., Shen, H. M., & Liou, Y. C. (2015). Death-associated protein 3 regulates mitochondrial-encoded protein synthesis and mitochondrial dynamics. Journal of Biological Chemistry, 290(41), 24961–24974. https://doi.org/10.1074/jbc.M115.673343.
Yasumura, K., Sugimura, I., Igarashi, K., Kakinuma, S., Nishimura, M., Doi, M., & Shimada, Y. (2004). Altered expression of tfg and Dap3 in Ikaros-defective T-cell lymphomas induced by X-irradiation in B6C3F1 mice. British Journal of Haematology, 124(2), 179–185. https://doi.org/10.1046/j.1365-2141.2003.04768.x.
Yepuri, G., Ramirez, L. M., Theophall, G. G., Reverdatto, S. V., Quadri, N., Hasan, S. N., Bu, L., Thiagarajan, D., Wilson, R., Diez, R. L., Gugger, P. F., Mangar, K., Narula, N., Katz, S. D., Zhou, B., Li, H., Stotland, A. B., Gottlieb, R. A., Schmidt, A. M., Shekhtman, A., & Ramasamy, R. (2023). DIAPH1-MFN2 interaction regulates mitochondria-SR/ER contact and modulates ischemic/hypoxic stress. Nature Communications, 14(1), 6900. https://doi.org/10.1038/s41467-023-42521-x.
Yin, Y. J., Zhang, Y. H., Wang, Y., Jiang, H., Zhang, J. B., Liang, S., & Yuan, B. (2023). Ferulic acid ameliorates the quality of in vitro-aged bovine oocytes by suppressing oxidative stress and apoptosis. Aging (Albany NY), 15(21), 12497–12512. https://doi.org/10.18632/aging.205193.
Zhang, S., Zhang, Y., Feng, Y., Wu, J., Hu, Y., Lin, L., Xu, C., Chen, J., Tang, Z., Tian, H., & Chen, X. (2022). Biomineralized two-enzyme nanoparticles regulate tumor glycometabolism inducing tumor cell pyroptosis and robust antitumor immunotherapy. Advanced Materials, 34(50). https://doi.org/10.1002/adma.202206851.
Zhao, W., Zhang, L., Zhang, Y., Jiang, Z., Lu, H., Xie, Y., Han, W., Zhao, W., He, J., Shi, Z., Yang, H., Chen, J., Chen, S., Li, Z., Mao, J., Zhou, L., Gao, X., Li, W., Tan, G., Zhang, B., & Wang, Z. (2023). The CDK inhibitor AT7519 inhibits human glioblastoma cell growth by inducing apoptosis, pyroptosis and cell cycle arrest. Cell Death & Disease, 14(1). https://doi.org/10.1038/s41419-022-05528-8.
Zhu, M., Wang, J., & Zhou, R. (2022). Combination of metformin and oxaliplatin inhibits gastric cancer cell proliferation and induces apoptosis. Acta Biochimica Polonica, 69(2), 321–326. https://doi.org/10.18388/abp.2020_5750.
Acknowledgements
This study was supported by National Natural Science Foundation of China (31771534, and 32370741); Scientific Research Foundation of University of South China (211RJC002); The Special Funding for the Construction of Innovative Provinces in Hunan (2021SK4031).
Author information
Ting Cao, Xuling Luo and Binjiao Zheng contributed equally to this work.
Authors and Affiliations
Department of Gastroenterology and Hunan Provincial Clinical Research Center for Metabolic Associated Fatty Liver Disease, Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, The Affiliated Nanhua Hospital, University of South China, Hengyang, Hunan, 421001, China
Ting Cao, Xuling Luo, Yao Deng, Yu Zhang, Wenwen Xi, Meng Guo, Xuefeng Yang & Bin Lu
National Health Commission Key Laboratory of Birth Defect Research and Prevention, Hunan Provincial Maternal and Child Health Care Hospital, Changsha, Hunan, 410008, China
Ting Cao, Xuling Luo, Yao Deng, Yu Zhang, Meng Guo & Bin Lu
MOE Key Lab of Rare Pediatric Diseases, Hengyang, 421001, China
Ting Cao, Xuling Luo, Yao Deng, Yu Zhang, Meng Guo & Bin Lu
Protein Quality Control and Diseases Laboratory, Zhejiang Provincial Key Laboratory of Medical Genetics, and Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, Zhejiang, 325035, China
Binjiao Zheng
Nanchang University Queen Mary School, Nanchang, Jiangxi, 330031, China
Yuyan Li
Department of Burn and Plastic Surgery Department, The Second Affiliated Hospital of Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China
Wenwen Xi
Department of Orthopedics, Third Xiangya Hospital of Central South University, Changsha, Hunan, 410013, China
Zhiyue Li
Corresponding authors
Correspondence to Xuefeng Yang, Zhiyue Li or Bin Lu.
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, T., Luo, X., Zheng, B. et al. Death-associated protein 3 in cell death and beyond. GENOME INSTAB. DIS. (2024). https://doi.org/10.1007/s42764-024-00125-9
Received01 December 2023
Revised19 February 2024
Accepted25 February 2024
Published04 March 2024
DOIhttps://doi.org/10.1007/s42764-024-00125-9
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable link







用户登录
还没有账号?
立即注册