KIF18A as a potential biomarker to distinguish different breast cancer subtypes based on receptor status
Original Research Paper
Published: 15 March 2024
Caglar Berkel
Genome Instability & Disease Volume 5, pages 89–96, (2024)
Abstract
The inhibition of KIF18A selectively reduces the viability of chromosomally unstable cancers due to increased mitotic vulnerability. KIF18A expression was also reported to be upregulated and associated with tumor aggressiveness in certain cancer types including breast cancer. Here, I first showed that KIF18A mRNA expression is higher in triple-negative breast cancer (TNBC) than in non-TNBC. I also found that ER (estrogen receptor)-negative and PR (progesterone receptor)-negative breast cancer cells have higher KIF18A mRNA expression compared to ER-positive and PR-positive breast cancer cells, respectively. In contrast, HER2-positive breast tumors have higher KIF18A expression compared to HER2-negative breast tumors. In terms of PAM50 breast cancer subtypes, KIF18A transcript levels were found to be the highest in basal-like breast cancer, followed by HER2-enriched, luminal B, normal-like and luminal A. Besides, in non-TNBC, cells with high AR (androgen receptor) mRNA expression have higher KIF18A mRNA expression than cells with low AR mRNA expression. Both non-TNBC and TNBC cells with high BRCA1 and BRCA2 mRNA expression levels were observed to have higher KIF18A mRNA expression than those with low BRCA1 and BRCA2 mRNA expression levels, respectively. Combined, this study demonstrates that breast tumors with low and high expression of ER, PR, HER2, AR and BRCA1/2 have differential transcript levels of KIF18A, pointing that KIF18A might contribute to the molecular differences between different breast cancer subtypes.
Introduction
KIF18A (Kinesin Family Member 18A), a plus-end directed microtubule depolymerase kinesin which regulates microtubule dynamics, chromosome congression and cell division by facilitating proper alignment and distribution of chromosomes during mitosis (Fonseca et al., 2019; Mayr et al., 2007; Stumpff et al., 2008, 2012), is overexpressed in breast cancer, and its overexpression was shown to be associated with tumor grade, metastasis and poor survival, and to be a predictive biomarker of poor benefit from endocrine therapy in early ER (estrogen receptor)-positive breast cancer (Alfarsi et al., 2019; Hitti et al., 2016; Kasahara et al., 2016; Li et al., 2020; Savci-Heijink et al., 2019; Zhang et al., 2010). Ectopic overexpression of KIF18A results in cell multinucleation, whereas ablation of KIF18A expression significantly inhibits breast cancer cell proliferation both in vitro and in vivo (Zhang et al., 2010). The depletion of KIF18A genetically leads to increased chromosome oscillation and elongated mitotic spindles in cell lines (Fonseca et al., 2019; Mayr et al., 2007; Stumpff et al., 2008). The inhibition of KIF18A, besides affecting its critical mitotic function, also decreases cancer cell migration by stabilizing microtubules at leading edges (Zhang et al., 2010). Furthermore, Quinton et al. (2021) showed that whole-genome doubling which generates genetically unstable tetraploid cells confers dependence on KIF18A in a panel of breast cancer cell lines. In addition, estrogen was shown to strongly induce the expression of KIF18A in ER-positive breast cancer cells (Zou et al., 2014).
Chromosomal instability is caused by persistent errors in chromosome segregation during mitosis, for instance, by alterations in the dynamics and control of mitotic spindle microtubules which compromise the mitotic spindle (Marquis et al., 2021; Payton et al., 2023). In recent years, it was shown that chromosomally unstable tumor cells (such as chromosomally unstable triple-negative breast cancer (TNBC) cell lines) specifically require KIF18A for their proliferation, and that KIF18A depletion increases cell death in these cells by inducing a prolonged mitotic delay and causing mitotic arrest and centrosome fragmentation (Marquis et al., 2021). In contrast, this protein is not required for the proliferation of near-diploid cells (Marquis et al., 2021). Genetic perturbation of KIF18A was shown to selectively reduce the viability of cancer cell lines with chromosomal instability, by inducing notable mitotic errors such as mitotic delays and multipolar spindles (Cohen-Sharir et al., 2021; Marquis et al., 2021; Quinton et al., 2021), and the inhibitors of the KIF18A motor protein were reported to activate the mitotic checkpoint and selectively kill chromosomally unstable cancer cells which depend on KIF18A to maintain bipolar spindle integrity (Payton et al., 2023). Sensitivity to KIF18A inhibition was found to be enriched in TNBC cell lines with chromosomal instability (Payton et al., 2023).
In the current study, I found that KIF18A mRNA expression is higher in triple-negative breast cancer (TNBC) than in non-TNBC. In parallel, I showed that, in non-TNBC, ER-negative and PR-negative breast tumors have higher KIF18A mRNA expression compared to ER-positive and PR-positive breast tumors, respectively. In contrast, I reported that HER2-positive breast cancer cells have higher KIF18A expression compared to HER2-negative breast cancer cells. Besides, I found that, in non-TNBC, cells with high AR (androgen receptor) mRNA expression have higher KIF18A mRNA expression than cells with low AR mRNA expression. Furthermore, both non-TNBC and TNBC cells with high BRCA1 and BRCA2 mRNA expression levels had higher KIF18A mRNA expression than those with low BRCA1 and BRCA2 mRNA expression levels, respectively. Combined, this study demonstrates that KIF18A mRNA expression might depend on ER, PR, HER2 and AR status, and might be associated with BRCA1/2 expression in breast cancer.
Methods
Datasets
In this study, TCGA-BRCA transcriptomics dataset (https://portal.gdc.cancer.gov/projects/TCGA-BRCA) was used, which was accessed through ExperimentHub and AnnotationHub Bioconductor packages as previously reported (Morgan & Shepherd, 2022a, b; Berkel, 2023; Berkel & Cacan, 2023a). Using query() function, the transcriptomics dataset with GEO accession ID of GSE62944 (TCGA re-processed RNA-Seq data) was loaded into R programming environment. These data have been parsed into SummarizedExperiment objects and are available in ExperimentHub package (2022b; Morgan & Shepherd, 2022a). In this dataset, ER, PR and HER2 status (positivity/negativity) were determined by IHC (immunohistochemistry). Patients samples only for which data for ER, PR and HER2 status are available were included in further analyses. Sample size (n) was decreased from 1119 to 703 after this filtering step. The distribution of breast cancer patients based on receptor status (by IHC) is as following: ER-negative = 160; ER-positive = 543; PR-negative = 233; PR-positive = 470; HER2-negative = 542; HER2-positive = 161. Of these patients, 591 patients have non-TNBC, and 112 patients have TNBC (triple-negative breast cancer). ESR1 (encoding ER-α), ESR2 (encoding ER-β), PGR, ERBB2 (encoding HER2), AR, BRCA1 and BRCA2 gene expression status (low vs high) by RNA-Seq were determined based on the median expression value for each gene. For instance, patients with higher than the median ESR1 mRNA expression were considered as patients with high ESR1 expression; and patients with lower than the median ESR1 mRNA expression were considered as patients with low ESR1 expression.
Data analysis and visualization
Data analysis and visualization in this study was completely performed in R programming environment as previously reported (R Core Team, 2022; Berkel & Cacan, 2021, 2023b). Following R and Bioconductor packages were used throughout the analysis: tidyverse (Wickham et al., 2019), ggpubr (Kassambara, 2023), magick (Ooms, 2023) and SummarizedExperiment (Morgan et al., 2022). The mRNA expression values were shown in log10 scale on the y-axis of each plot. Means of expression values for each group were shown in red on the boxplots. The lower and upper hinges of boxplots correspond to the first and third quartiles (the 25th and 75th percentiles) (Wickham et al., 2019). The normality of the data was tested using the Shapiro–Wilk test of normality (Royston, 1995). When the resultant p < 0.05, we could not assume normality, and therefore performed wilcox test (otherwise, t test was used to compare group means).
Results
KIF18A mRNA expression is higher in TNBC than in non-TNBC
I first showed that KIF18A transcript levels are higher in tumors from TNBC (triple-negative breast cancer) patients than in tumors from non-TNBC patients (Fig. 1, p < 2e−16). This observation supports recent findings showing that sensitivity to KIF18A inhibition is enriched in TNBC cell lines with chromosomal instability features, and that KIF18A is required for the proliferation of chromosomally instable cells derived from TNBC (Marquis et al., 2021; Payton et al., 2023).
Fig. 1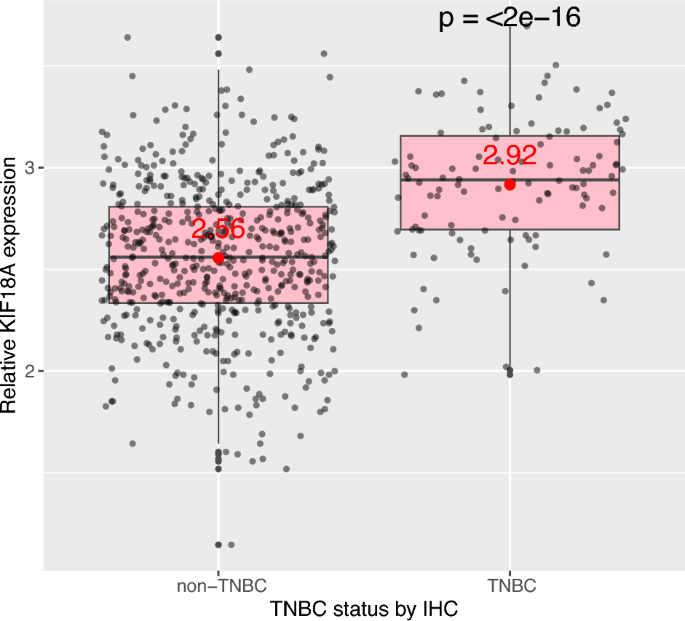
KIF18A mRNA expression is higher in TNBC than in non-TNBC. Relative KIF18A transcript levels between tumors from patients with TNBC or non-TNBC. ER-, PR- and HER2-status were determined by IHC. TNBC: Triple negative breast cancer (ER-negative, PR-negative, HER2-negative); IHC: Immunohistochemistry. Sample size (n) for patients with non-TNBC = 591, and for patients with TNBC (triple-negative breast cancer) = 112. Data from TCGA-BRCA (https://portal.gdc.cancer.gov/projects/TCGA-BRCA), obtained programmatically using SummarizedExperiment Bioconductor package (https://bioconductor.org/packages/release/bioc/html/SummarizedExperiment.html). Mean expression values were given in red
ER-negative and PR-negative breast cancer cells have higher KIF18A mRNA expression compared to ER-positive and PR-positive breast cancer cells, respectively
In parallel to data shown in Fig. 1, I found that, even within patients with non-TNBC, ER (estrogen receptor)-negative and PR (progesterone)-negative breast tumors have higher KIF18A mRNA expression compared to ER-positive and PR-positive breast tumors, respectively (Fig. 2, first two plots, p values = 0.00037 and 0.0087, respectively). However, this difference at KIF18A transcript levels was only observed when ER and PR status were determined at the protein level (at the translational level) (i.e. when ER and PR levels/status were determined by IHC (immunohistochemistry)) (Fig. 2, first two plots), but not at the mRNA level (i.e. when ER-α (ESR1), ER-β (ESR2), and PR (PGR1) mRNA levels were determined by RNA-Seq) (Fig. 3, first three plots). When ER and PR levels were determined by RNA-Seq (i.e. at the transcription level in contrast to the translational level), I found that non-TNBC cells with high ESR2 (encoding ER-β) mRNA expression have slightly higher KIF18A expression than non-TNBC cells with low ESR2 mRNA expression (Fig. 3, second plot, p = 0.033), and that non-TNBC cells with low PGR (encoding PR) mRNA expression have slightly higher KIF18A expression than non-TNBC cells with high PGR mRNA expression (Fig. 3, third plot, p = 0.051).
Fig. 2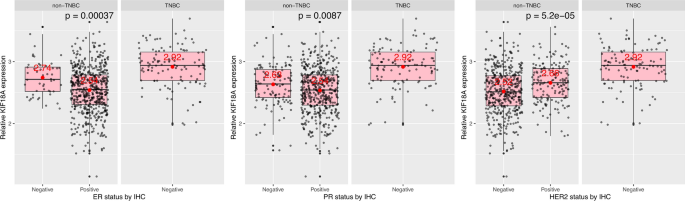
ER-negative and PR-negative breast cancer cells have higher KIF18A mRNA expression compared to ER-positive and PR-positive breast cancer cells, respectively. Comparative KIF18A transcript levels between tumors of negative and positive status for ER, PR or HER2 in patients with non-TNBC. ER-, PR- and HER2-status were determined by IHC. ER: Estrogen receptor; PR: Progesterone receptor; HER2: Human epidermal growth factor receptor-2; IHC: Immunohistochemistry; TNBC: Triple negative breast cancer (ER-negative, PR-negative, HER2-negative). The distribution of breast cancer patients based on receptor status (by IHC) is as following: ER-negative = 160; ER-positive = 543; PR-negative = 233; PR-positive = 470; HER2-negative = 542; HER2-positive = 161. Data from TCGA-BRCA, obtained programmatically using SummarizedExperiment Bioconductor package (https://bioconductor.org/packages/release/bioc/html/SummarizedExperiment.html). Mean expression values were given in red
The influence of ESR1, ESR2, PGR and ERBB2 mRNA expression on KIF18A mRNA expression in TNBC and non-TNBC. ESR1: Estrogen Receptor 1, encoding ER-α; ESR2: Estrogen Receptor 2, encoding ER-β; PGR: Progesterone Receptor, encoding PR; ERBB2: Erb-B2 Receptor Tyrosine Kinase 2, encoding HER2; IHC: Immunohistochemistry; TNBC: Triple negative breast cancer (ER-negative, PR-negative, HER2-negative); RNA-Seq: RNA sequencing. Data from TCGA-BRCA, obtained programmatically using SummarizedExperiment Bioconductor package (https://bioconductor.org/packages/release/bioc/html/SummarizedExperiment.html). Mean expression values were given in red
HER2-positive breast cancer cells have higher KIF18A expression compared to HER2-negative breast cancer cells
Next, I found that, in non-TNBC, in contrast to what was observed for ER and PR status, HER2-positive breast tumors cells have higher KIF18A transcript levels compared to HER2-negative breast tumors (Fig. 2, last plot, p = 5.2e−05). This observation was only valid when HER2 status was determined at the protein level (by IHC), since breast tumors with low and high HER2 mRNA expression have similar levels of KIF18A mRNA expression (Fig. 3, last plot). Combined, it can be stated that high KIF18A mRNA expression in TNBC (compared to non-TNBC) is mainly due to hormone receptor (ER and PR) status, not HER2 status. I also found that the influence of KIF18A expression on overall patient survival (OS) is similar between patients with all subtypes combined and HER2-negative breast cancer patients (hazard ratios (HRs) of 1.85 and 1.82, respectively) (Supplementary Fig. 1; for other breast cancer subtypes, FDR (false discovery rate) for Kaplan Meier curves is higher than 1%; therefore, they are not included / shown).
Similar changes in the mRNA expression of KIF18A based on ER-, PR- and HER2-status in breast tumors collected from patients were observed using another transcriptomics dataset (the one from METABRIC (Molecular Taxonomy of Breast Cancer International Consortium), accessed via https://www.mercuriolab.umassmed.edu/metabric (Curtis et al., 2012)) (Supplementary Fig. 2).
PAM50 is a 50-gene signature that classifies breast cancer into five molecular intrinsic subtypes: Luminal A, Luminal B, HER2-enriched, Basal-like and Normal-like. In terms of PAM50 breast cancer subtypes, KIF18A expression is the highest in Basal-like breast cancer, followed by HER2-enriched, Luminal B, Normal-like and Luminal A (Fig. 4) (Goldman et al., 2020). Please note that Luminal A subtype generally has the best prognosis; and Basal-like and HER2-enriched breast cancers are considered more aggressive diseases (Kensler et al., 2019). In other words, KIF18A mRNA expression is higher in more aggressive PAM50 subtypes of breast cancer (i.e. Basal-like and HER2-enriched), similar to what I observed in terms of triple-negativity status (higher expression of KIF18A in TNBC which tends to have a worse prognosis).
Fig. 4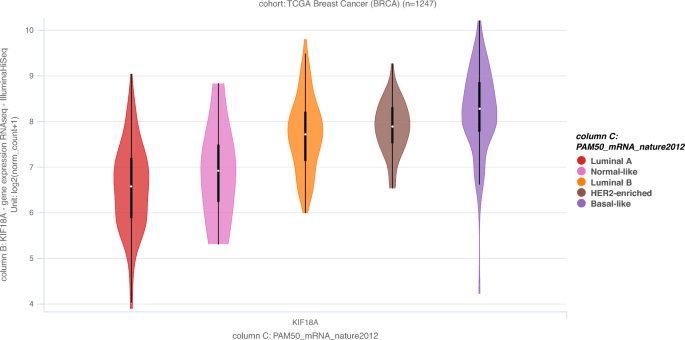
KIF18A mRNA expression based on PAM50 breast cancer subtypes. Sample size (n) = 1247. Data from TCGA-BRCA obtained via Xena Functional Genomics Explorer (https://xenabrowser.net/) (Goldman et al., 2020)
Androgen receptor (AR) mRNA expression-, BRCA1 mRNA expression- and BRCA2 mRNA expression-dependent changes in the KIF18A transcript levels in non-TNBC and TNBC
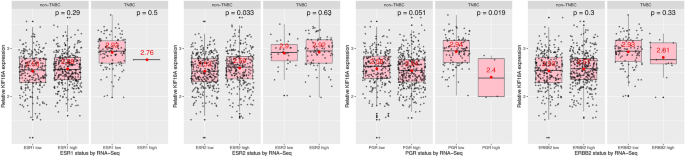
Lastly, I studied KIF18A mRNA expression based on AR (androgen receptor), BRCA1 and BRCA2 expression levels (low vs high) which was determined by RNA-Seq (Fig. 5). I found that, in non-TNBC, cells with high AR mRNA expression have higher KIF18A transcript levels than cells with low AR mRNA expression (Fig. 5, first plot, first subplot, p = 0.0025). In contrast, there was no significant change in KIF18A mRNA expression in TNBC cells with low and high AR mRNA expression (Fig. 5, first plot, second subplot, p = 0.062). In terms of BRCA1 and BRCA2 mRNA expression levels, both non-TNBC and TNBC cells with high BRCA1 and BRCA2 mRNA expression levels have higher KIF18A transcript levels than those with low BRCA1 and BRCA2 mRNA expression levels, respectively (Fig. 5, last two plots, p values = < 2e−16, 9e−04, < 2e−16 and 0.00097, respectively). In other words, KIF18A mRNA expression is correlated with BRCA1/2 mRNA expression in both non-TNBC and TNBC cells (Fig. 5, last two plots).
Fig. 5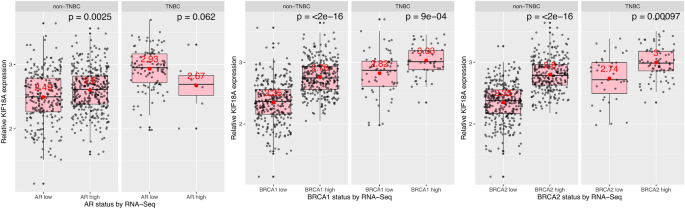
Androgen receptor (AR) mRNA expression-, BRCA1 mRNA expression- and BRCA2 mRNA expression-dependent changes in the KIF18A expression in non-TNBC and TNBC. TNBC: Triple negative breast cancer (ER-negative, PR-negative, HER2-negative); RNA-Seq: RNA sequencing. Data from TCGA-BRCA, obtained programmatically using SummarizedExperiment Bioconductor package (https://bioconductor.org/packages/release/bioc/html/SummarizedExperiment.html). Mean expression values were given in red
In addition to patient tumor data shown above, breast cancer cell line data (namely, KIF18A protein levels based on breast cancer subtypes (HER2 Amp, Basal A, Basal B and Luminal), KIF18A mRNA levels based on BRCA1/2 damaging mutation status and certain subtypes (Luminal HER2 Amp, Luminal, Basal, Basal A, Basal B, HER2 Amp, ER+ HER2+ , ER+ HER2−, ER−HER2+, ER−HER2−)) was also given in Supplementary Fig. 3.
Discussion
It was recently reported that the sensitivity to KIF18A inhibition is enriched in TNBC cell lines with chromosomal instability (Payton et al., 2023). In the present study, I found that breast tumors with triple-negative status (ER−, PR− and HER2−) have higher KIF18A transcript levels than those with no triple-negative status (non-TNBC). KIF18A is elevated in actively proliferating tissues and in certain cancer types including breast cancer; and it is associated with tumor aggressiveness (Mayr et al., 2007; Rath & Kozielski, 2012). Besides, since chromosomally unstable cancer cells specifically require KIF18A for their proliferation (Marquis et al., 2021), cells with high KIF18A expression might be positively selected during cancer evolution from within TNBC cells which are characterized by high levels of genetic instability (Derakhshan & Reis-Filho, 2022; Pareja et al., 2013; Turner & Reis-Filho, 2013), possibly resulting in the higher expression of KIF18A observed in TNBC than in non-TNBC. Relatively high KIF18A expression in TNBC might also contribute to the most aggressive behavior and unfavorable prognosis in TNBC of all breast cancer subtypes (for instance, compared to non-TNBC) (Tečić Vuger et al., 2020). In other words, increased KIF18A levels in TNBC might be associated, at least to a certain extent, with high tumor grade and increased number of metastatic events. In parallel, I also observed that, in non-TNBC, ER-negative and PR-negative breast tumors have increased mRNA expression of KIF18A compared to those with ER-negative and PR-negative status, respectively. In other words, ER or PR negativity was associated with higher KIF18A mRNA expression in non-TNBC. It can be speculated that some proteins whose expression was positively regulated (up-regulated) by ER and/or PR might negatively regulate KIF18A expression in breast tumors, leading to decreased expression of KIF18A in ER+ and PR+ breast tumors compared to those with ER− and PR− status, respectively. Considering the fact that patients with ER+ breast cancer have a better prognosis than patients with ER− breast cancer (Belete et al., 2022), decreased expression of KIF18A in ER+ breast cancer might lead to less agressive tumor behaviour, ultimately resulting in more favorable prognosis in this patient group. Besides, estrogen induces the expression of KIF18A in ER-positive breast cancer cells (Zou et al., 2014); therefore, higher expression of KIF18A in ER-negative breast cancer cells might be a compensatory mechanism for decreased estrogen responsiveness (due to lack of ERs). In contrast to ER and PR, I found that HER2+ breast tumors have higher KIF18A expression than breast tumors with HER2− status. Since HER2 positivity is associated with higher rates of disease recurrence and mortality (in contrast to ER+) (Patel et al., 2020), increased expression of KIF18A in HER2+ breast tumors might contribute to unfavorable prognosis observed in patients with HER2+ breast tumors.
Breast tumors with BRCA gene mutations often display a basal phenotype and are triple-negative (Greenup et al., 2013; Lal et al., 2019;), and BRCA mutation carriers have significantly worse survival when compared with non-carriers (Huszno et al., 2019). Since I observed that KIF18A expression is higher in basal-like breast cancer and in TNBC (subtypes with unfavorable prognosis), BRCA mutation carriers might similarly have increased KIF18A expression, again contributing to poorer outcomes in this patient group.
Androgen receptor (AR) is a steroid hormone receptor expressed in around 70% of breast cancers (Ravaioli et al., 2022). Previous research suggested that AR might be a tumor suppressor in ER alpha-positive breast cancer; but, a tumor promoter in ER alpha-negative breast cancer (You et al., 2022). I found that, in non-TNBC (but not in TNBC), AR-high tumors have higher KIF18A transcript levels compared to AR-low tumors, pointing that AR status might differentialy influence or be associated with KIF18A expression in non-TNBC. Lastly, the analysis demonstrated that BRCA1- or BRCA2-high breast tumors have higher expression of KIF18A than BRCA1- or BRCA2-low breast tumors, respectively, both in non-TNBC and TNBC. In other words, BRCA1/2 high expression might be associated with high KIF18A expression in breast cancer independent of ER, PR and HER2 status. Since high BRCA1/2 expression is known to be associated with poor prognosis in breast cancer (Chang et al., 2022; Jin et al., 2022; Wang et al., 2018), it can be proposed that increased KIF18A expression observed in breast tumors from patients with high BRCA1/2 expression might contribute to unfavorable outcomes in these patients, at least to a certain level. It can also be suggested that since both BRCA1/2 and KIF18A participate in the maintenance of genome stability (Gorodetska et al., 2019; Gudmundsdottir & Ashworth, 2006), their expression might possibly be regulated by parallel upstream mechanisms, leading to the coordinated upregulation of their expression, potentially contributing to increased genome stability. However, further research is required to test these assumptions.
Combined, the present study demonstrated that KIF18A expression in breast cancer is influenced by the ER, PR, HER2, AR and BRCA1/2 expression status, and that its expression is elevated in TNBC compared to non-TNBC, possibly contributing to unfavorable prognosis in TNBC due to the association of high KIF18A levels with high tumor grade and metastasis in breast cancer.
Data availability
Data used in the present study is publicly available, for which resources were given in the Methods section.
References
Alfarsi, L. H., Elansari, R., Toss, M. S., Diez-Rodriguez, M., Nolan, C. C., Ellis, I. O., Rakha, E. A., & Green, A. R. (2019). Kinesin family member-18A (KIF18A) is a predictive biomarker of poor benefit from endocrine therapy in early ER+ breast cancer. Breast Cancer Research and Treatment, 173(1), 93–102. https://doi.org/10.1007/s10549-018-4978-5. Epub 2018 Oct 10. PMID: 30306428.
Belete, A. M., Aynalem, Y. A., Gemeda, B. N., Demelew, T. M., & Shiferaw, W. S. (2022). The effect of estrogen receptor status on survival in breast cancer patients in Ethiopia. Retrospective Cohort Study. Breast Cancer (Dove Med Press), 14, 153–161. https://doi.org/10.2147/BCTT.S365295.
Berkel, C. (2023). Estrogen receptor- and progesterone receptor-positive breast tumors have higher mRNA levels of NR3C1 and ZBTB16, with implications in prognosis for luminal A subtype. Human Cell. https://doi.org/10.1007/s13577-023-01014-1. Epub ahead of print. PMID: 37999919.
Berkel, C., & Cacan, E. (2021). Involvement of ATMIN-DYNLL1-MRN axis in the progression and aggressiveness of serous ovarian cancer. Biochemical and Biophysical Research Communications, 17(570), 74–81. https://doi.org/10.1016/j.bbrc.2021.07.004. Epub 2021 Jul 14. PMID: 34273621.
Berkel, C., & Cacan, E. (2023a). Lower expression of NINJ1 (Ninjurin 1), a mediator of plasma membrane rupture, is associated with advanced disease and worse prognosis in serous ovarian cancer. Immunologic Research, 71(1), 15–28. https://doi.org/10.1007/s12026-022-09323-7. Epub 2022 Oct 3. PMID: 36184655.
Berkel, C., & Cacan, E. (2023b). The expression of O-linked glycosyltransferase GALNT7 in breast cancer is dependent on estrogen-, progesterone-, and HER2-receptor status, with prognostic implications. Glycoconjugate Journal. https://doi.org/10.1007/s10719-023-10137-4. Epub ahead of print. PMID: 37947928.
Chang, H. J., Yang, U. C., Lai, M. Y., Chen, C. H., & Fann, Y. C. (2022). High BRCA1 gene expression increases the risk of early distant metastasis in ER+ breast cancers. Science and Reports, 12(1), 77. https://doi.org/10.1038/s41598-021-03471-w. PMID: 34996912; PMCID: PMC8741892.
Cohen-Sharir, Y., McFarland, J. M., Abdusamad, M., Marquis, C., Bernhard, S. V., Kazachkova, M., Tang, H., Ippolito, M. R., Laue, K., Zerbib, J., Malaby, H. L. H., Jones, A., Stautmeister, L. M., Bockaj, I., Wardenaar, R., Lyons, N., Nagaraja, A., Bass, A. J., Spierings, D. C. J., … Ben-David, U. (2021). Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature, 590(7846), 486–491. https://doi.org/10.1038/s41586-020-03114-6. Epub 2021 Jan 27. PMID: 33505028; PMCID: PMC8262644.
Curtis, C., Shah, S. P., Chin, S. F., Turashvili, G., Rueda, O. M., Dunning, M. J., Speed, D., Lynch, A. G., Samarajiwa, S., Yuan, Y., Gräf, S., Ha, G., Haffari, G., Bashashati, A., Russell, R., McKinney, S., METABRIC Group, Langerød, A., Green, A., … Aparicio, S. (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature, 486(7403), 346–352. https://doi.org/10.1038/nature10983. PMID: 22522925; PMCID: PMC3440846.
Derakhshan, F., & Reis-Filho, J. S. (2022). Pathogenesis of triple-negative breast cancer. Annual Review of Pathology: Mechanisms of Disease, 24(17), 181–204. https://doi.org/10.1146/annurev-pathol-042420-093238. PMID: 35073169; PMCID: PMC9231507.
Fonseca, C. L., Malaby, H. L. H., Sepaniac, L. A., Martin, W., Byers, C., Czechanski, A., Messinger, D., Tang, M., Ohi, R., Reinholdt, L. G., & Stumpff, J. (2019). Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase. Journal of Cell Biology, 218(4), 1148–1163. https://doi.org/10.1083/jcb.201807228. Epub 2019 Feb 7. PMID: 30733233; PMCID: PMC6446859.
Goldman, M. J., Craft, B., Hastie, M., Repečka, K., McDade, F., Kamath, A., Banerjee, A., Luo, Y., Rogers, D., Brooks, A. N., Zhu, J., & Haussler, D. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology, 38(6), 675–678. https://doi.org/10.1038/s41587-020-0546-8. PMID: 32444850; PMCID: PMC7386072.
Gorodetska, I., Kozeretska, I., & Dubrovska, A. (2019). BRCA genes: the role in genome stability, cancer stemness and therapy resistance. Journal of Cancer, 10(9), 2109–2127. https://doi.org/10.7150/jca.30410. PMID: 31205572; PMCID: PMC6548160.
Greenup, R., Buchanan, A., Lorizio, W., Rhoads, K., Chan, S., Leedom, T., King, R., McLennan, J., Crawford, B., Kelly Marcom, P., & Shelley, H. E. (2013). Prevalence of BRCA mutations among women with triple-negative breast cancer (TNBC) in a genetic counseling cohort. Annals of Surgical Oncology, 20(10), 3254–3258. https://doi.org/10.1245/s10434-013-3205-1. Epub 2013 Aug 22. PMID: 23975317.
Gudmundsdottir, K., & Ashworth, A. (2006). The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene, 25(43), 5864–5874. https://doi.org/10.1038/sj.onc.1209874. PMID: 16998501.
Győrffy, B. (2021). Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Computational and Structural Biotechnology Journal, 18(19), 4101–4109. https://doi.org/10.1016/j.csbj.2021.07.014. PMID: 34527184; PMCID: PMC8339292.
Hitti, E., Bakheet, T., Al-Souhibani, N., Moghrabi, W., Al-Yahya, S., Al-Ghamdi, M., Al-Saif, M., Shoukri, M. M., Lánczky, A., Grépin, R., Győrffy, B., Pagès, G., & Khabar, K. S. (2016). Systematic analysis of AU-rich element expression in cancer reveals common functional clusters regulated by key RNA-binding proteins. Cancer Research, 76(14), 4068–4080. https://doi.org/10.1158/0008-5472.CAN-15-3110. Epub 2016 May 17. PMID: 27197193.
Huszno, J., Kołosza, Z., & Grzybowska, E. (2019). BRCA1 mutation in breast cancer patients: Analysis of prognostic factors and survival. Oncology Letters, 17(2), 1986–1995. https://doi.org/10.3892/ol.2018.9770. Epub 2018 Nov 28. PMID: 30675265; PMCID: PMC6341769.
Jin, T. Y., Park, K. S., Nam, S. E., Yoo, Y. B., Park, W. S., & Yun, I. J. (2022). BRCA1/2 serves as a biomarker for poor prognosis in breast carcinoma. International Journal of Molecular Sciences, 23(7), 3754. https://doi.org/10.3390/ijms23073754. PMID: 35409110; PMCID: PMC8998777.
Kasahara, M., Nagahara, M., Nakagawa, T., Ishikawa, T., Sato, T., Uetake, H., & Sugihara, K. (2016). Clinicopathological relevance of kinesin family member 18A expression in invasive breast cancer. Oncology Letters, 12(3), 1909–1914. https://doi.org/10.3892/ol.2016.4823. Epub 2016 Jul 7. PMID: 27588139; PMCID: PMC4998100.
Kassambara, A. (2023). ggpubr: 'ggplot2' based publication ready plots. R package version 0.6.0. https://CRAN.R-project.org/package=ggpubr. Accessed 26 Dec 2023.
Kensler, K. H., Sankar, V. N., Wang, J., Zhang, X., Rubadue, C. A., Baker, G. M., Parker, J. S., Hoadley, K. A., Stancu, A. L., Pyle, M. E., Collins, L. C., Hunter, D. J., Eliassen, A. H., Hankinson, S. E., Tamimi, R. M., & Heng, Y. J. (2019). PAM50 molecular intrinsic subtypes in the nurses’ health study cohorts. Cancer Epidemiology, Biomarkers & Prevention, 28(4), 798–806. https://doi.org/10.1158/1055-9965.EPI-18-0863. Epub 2018 Dec 27. PMID: 30591591; PMCID: PMC6449178.
Lal, A., Ramazzotti, D., Weng, Z., Liu, K., Ford, J. M., & Sidow, A. (2019). Comprehensive genomic characterization of breast tumors with BRCA1 and BRCA2 mutations. BMC Medical Genomics, 12(1), 84. https://doi.org/10.1186/s12920-019-0545-0. PMID: 31182087; PMCID: PMC6558765.
Li, T. F., Zeng, H. J., Shan, Z., Ye, R. Y., Cheang, T. Y., Zhang, Y. J., Lu, S. H., Zhang, Q., Shao, N., & Lin, Y. (2020). Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell International, 15(20), 123. https://doi.org/10.1186/s12935-020-01191-1. PMID: 32322170; PMCID: PMC7161125.
Marquis, C., Fonseca, C. L., Queen, K. A., Wood, L., Vandal, S. E., Malaby, H. L. H., Clayton, J. E., & Stumpff, J. (2021). Chromosomally unstable tumor cells specifically require KIF18A for proliferation. Nature Communications, 12(1), 1213. https://doi.org/10.1038/s41467-021-21447-2. PMID: 33619254; PMCID: PMC7900194.
Mayr, M. I., Hümmer, S., Bormann, J., Grüner, T., Adio, S., Woehlke, G., & Mayer, T. U. (2007). The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Current Biology, 17(6), 488–498. https://doi.org/10.1016/j.cub.2007.02.036. Epub 2007 Mar 8. PMID: 17346968.
Morgan, M., Obenchain, V., Hester, J., & Pagès, H. (2022). SummarizedExperiment: SummarizedExperiment container. R package version 1.26.1. https://bioconductor.org/packages/SummarizedExperiment. Accessed 26 Dec 2023.
Morgan, M., Shepherd, L. (2022a). AnnotationHub: Client to access AnnotationHub resources. R package version 3.4.0.
Morgan, M., Shepherd, L. (2022b). ExperimentHub: Client to access ExperimentHub resources. R package version 2.4.0.
Ooms, J. (2023). magick: Advanced graphics and image-processing in R. R package version 2.7.4. https://CRAN.R-project.org/package=magick. Accessed 26 Dec 2023.
Pareja, F., Geyer, F. C., Marchiò, C., Burke, K. A., Weigelt, B., & Reis-Filho, J. S. (2016). Triple-negative breast cancer: The importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer, 16(2), 16036. https://doi.org/10.1038/npjbcancer.2016.36. PMID: 28721389; PMCID: PMC5515338.
Patel, A., Unni, N., & Peng, Y. (2020). The Changing paradigm for the treatment of HER2-positive breast cancer. Cancers (basel), 12(8), 2081. https://doi.org/10.3390/cancers12082081. PMID: 32731409; PMCID: PMC7464074.
Payton, M., Belmontes, B., Hanestad, K., Moriguchi, J., Chen, K., McCarter, J. D., Chung, G., Ninniri, M. S., Sun, J., Manoukian, R., Chambers, S., Ho, S. M., Kurzeja, R. J. M., Edson, K. Z., Dahal, U. P., Wu, T., Wannberg, S., Beltran, P. J., Canon, J., … Hughes, P. E. (2023). Small-molecule inhibition of kinesin KIF18A reveals a mitotic vulnerability enriched in chromosomally unstable cancers. Nature Cancer. https://doi.org/10.1038/s43018-023-00699-5. Epub ahead of print. PMID: 38151625.
Quinton, R. J., DiDomizio, A., Vittoria, M. A., Kotýnková, K., Ticas, C. J., Patel, S., Koga, Y., Vakhshoorzadeh, J., Hermance, N., Kuroda, T. S., Parulekar, N., Taylor, A. M., Manning, A. L., Campbell, J. D., & Ganem, N. J. (2021). Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature, 590(7846), 492–497. https://doi.org/10.1038/s41586-020-03133-3. Epub 2021 Jan 27. Erratum in: Nature. 2021 May;593(7860):E15. PMID: 33505027; PMCID: PMC7889737.
R Core Team. (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
Rath, O., & Kozielski, F. (2012). Kinesins and cancer. Nature Reviews Cancer, 12(8), 527–539. https://doi.org/10.1038/nrc3310. PMID: 22825217.
Ravaioli, S., Maltoni, R., Pasculli, B., Parrella, P., Giudetti, A. M., Vergara, D., Tumedei, M. M., Pirini, F., & Bravaccini, S. (2022). Androgen receptor in breast cancer: the “5W” questions. Frontiers in Endocrinology (lausanne)., 30(13), 977331. https://doi.org/10.3389/fendo.2022.977331. PMID: 36111296; PMCID: PMC9468319.
Royston, P. (1995). Remark AS R94: a remark on algorithm AS 181: The WW test for normality. Journal of Applied Statistics, 44, 547–551. https://doi.org/10.2307/2986146
Savci-Heijink, C. D., Halfwerk, H., Koster, J., Horlings, H. M., & van de Vijver, M. J. (2019). A specific gene expression signature for visceral organ metastasis in breast cancer. BMC Cancer, 19(1), 333. https://doi.org/10.1186/s12885-019-5554-z. PMID: 30961553; PMCID: PMC6454625.
Stumpff, J., von Dassow, G., Wagenbach, M., Asbury, C., & Wordeman, L. (2008). The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Developmental Cell, 14(2), 252–262. https://doi.org/10.1016/j.devcel.2007.11.014. PMID: 18267093; PMCID: PMC2267861.
Stumpff, J., Wagenbach, M., Franck, A., Asbury, C. L., & Wordeman, L. (2012). Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Developmental Cell, 22(5), 1017–1029. https://doi.org/10.1016/j.devcel.2012.02.013. PMID: 22595673; PMCID: PMC3356572.
TečićVuger, A., Šeparović, R., Vazdar, L., Pavlović, M., Lepetić, P., Šitić, S., Bajić, Ž, Šarčević, B., & Vrbanec, D. (2020). Characteristics and prognosis of triple-negative breast cancer patients: a croatian single institution retrospective cohort study. Acta Clinica Croatica, 59(1), 97–108. https://doi.org/10.20471/acc.2020.59.01.12. PMID: 32724280; PMCID: PMC7382886.
Turner, N. C., & Reis-Filho, J. S. (2013). Tackling the diversity of triple-negative breast cancer. Clinical Cancer Research, 19(23), 6380–6388. https://doi.org/10.1158/1078-0432. CCR-13-0915. PMID: 24298068.
Wang, Z., Zhang, J., Zhang, Y., Deng, Q., & Liang, H. (2018). Expression and mutations of BRCA in breast cancer and ovarian cancer: Evidence from bioinformatics analyses. International Journal of Molecular Medicine, 42(6), 3542–3550. https://doi.org/10.3892/ijmm.2018.3870. Epub 2018 Sep 11. PMID: 30221688.
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L. D., François, R., Grolemund, G., Hayes, A., Henry, L., Hester, J., Kuhn, M., Pedersen, T. L., Miller, E., Bache, S. M., Müller, K., Ooms, J., Robinson, D., Seidel, D. P., Spinu, V., … Yutani, H. (2019). Welcome to the tidyverse. Journal of Open Source Software, 4(43), 1686. https://doi.org/10.21105/joss.01686
You, C. P., Tsoi, H., Man, E. P. S., Leung, M. H., & Khoo, U. S. (2022). Modulating the activity of androgen receptor for treating breast cancer. International Journal of Molecular Sciences, 23(23), 15342. https://doi.org/10.3390/ijms232315342. PMID: 36499670; PMCID: PMC9739178.
Zhang, C., Zhu, C., Chen, H., Li, L., Guo, L., Jiang, W., & Lu, S. H. (2010). Kif18A is involved in human breast carcinogenesis. Carcinogenesis, 31(9), 1676–1684. https://doi.org/10.1093/carcin/bgq134. Epub 2010 Jul 1. PMID: 20595236.
Zou, J. X., Duan, Z., Wang, J., Sokolov, A., Xu, J., Chen, C. Z., Li, J. J., & Chen, H. W. (2014). Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Molecular Cancer Research, 12(4), 539–549. https://doi.org/10.1158/1541-7786.MCR-13-0459. Epub 2014 Jan 3. PMID: 24391143; PMCID: PMC4139106.
Author information
Authors and Affiliations
Department of Molecular Biology and Genetics, Tokat Gaziosmanpasa University, Tokat, Turkey
Caglar Berkel
Corresponding author
Correspondence to Caglar Berkel.
Ethics declarations
Conflict of interest
The author declares no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
42764_2024_126_MOESM1_ESM.pdf
Supplementary Figure 1. Kaplan-Meier curves showing overall survival (OS) for breast cancer patients with low and high expression of KIF18A. Left: all subtypes combined. Right: HER2-negative. HR: Hazard ratio. Data from [Győrffy, 2021] (PDF 971 KB)
42764_2024_126_MOESM2_ESM.pdf
Supplementary Figure 2 KIF18A mRNA expression levels based on ER-, PR- and HER2 status analysed using data from another dataset to validate findings from TCGA dataset. Data from METABRIC (Molecular Taxonomy of Breast Cancer International Consortium), accessed via https://www.mercuriolab.umassmed.edu/metabric [Curtis et al., 2012] (PDF 3930 KB)
42764_2024_126_MOESM3_ESM.pdf
Supplementary Figure 3 KIF18A mRNA and protein levels from cell lines based on breast cancer subtypes and BRCA1/2 mutation status. Data analysed using Dependency Map (DepMap) portal (https://depmap.org/portal/interactive/) (PDF 219 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Berkel, C. KIF18A as a potential biomarker to distinguish different breast cancer subtypes based on receptor status. GENOME INSTAB. DIS. 5, 89–96 (2024). https://doi.org/10.1007/s42764-024-00126-8
Received15 January 2024
Revised16 February 2024
Accepted25 February 2024
Published15 March 2024
Issue DateApril 2024
DOIhttps://doi.org/10.1007/s42764-024-00126-8
Share this article
Anyone you share the following link with will be able to read this content:
Get shareable link







用户登录
还没有账号?
立即注册