Dual roles of UFMylation on stalling fork stability
Published: 28 May 2024
Yisui Xia, Wenpeng Liu & Huiqiang Lou
Volume 5, pages 127–130, (2024)
Abstract
The mechanism governing the stabilization of the replication fork under replication stress is pivotal for maintaining genomic integrity and cellular viability. In this context, the safeguard factors BRCA1/2 and nucleases engage in a regulatory equilibrium, modulating the extent of nascent strand end resection—a process vital for replication fork stabilization under stress and for fork restart upon stress released. The recruitment dynamics of these nucleases, however, remain to be elucidated. Recent two independent studies by Gong et al. and Tian et al. have demonstrated that ubiquitin-like modification UFMylation employs dual pathways to facilitate the recruitment of nuclease MRE11, integral to the fork reversal mechanism. These revelations uncover novel roles of UFMylation in genome stability and provide guidance in identifying novel targets for treating BRCA1/2-mutated tumors.
Main text
DNA replication fidelity and genome stability, central to biological inheritance, are constantly challenged by transcription interference, DNA secondary structures, DNA protein crosslinks, etc., collectively termed replication stresses. Cells have evolved intricate mechanisms to rectify these issues, ensuring genome stability (Berti et al., 2020). Upon replication stress, DNA polymerases uncoupled from replicative helicase CMG, leading to single-strand DNA gap accumulation between helicase and polymerases (Lopes et al., 2006). Single-strand DNA binding protein RPA complex, which can be a platform to recruit RAD9-HUS1-RAD1 (9-1-1), ATRIP, TopBP1, and ATR to activate replication stress response mechanisms that preserve the integrity of the arrested replication fork and ensure genomic stability (Saldivar et al., 2017; Cortez, 2019; Berti et al., 2020). Fork reversal is a global response to many types of replication stress. Fork reversal indicates nascent DNA strands are reannealed together, accompanied by replication fork move backward (Zellweger et al., 2015).
Fork reversal is speculated to slow down the replication speed under stress conditions, facilitating the repair of lesions and recoupling of leading and lagging strands by template switching mechanisms (Thakar et al., 2021). Although CMG unloading/collapse from a stalled fork is the prerequisite for fork reversal in Xenopus egg extract (Amunugama et al., 2018), this suggests fork reversal might be a dead-end event since CMG is required for stalled fork restart once the stress is removed. Recent studies showed the compatibility of fork reversal and CMG at stalled forks in human cells. Fork reversal can occur by trapping CMG behind rather than unloading it for fork restart (Liu et al., 2023; Kavlashvili et al., 2023). On the other hand, reversed forks as intermediates might be toxic. The reversed arm is an ended double-strand DNA that tends to be degraded by multiple nucleases, such as MRE11, DNA2, EXO1, and MUS81. Although controlled resection contributes to stalled fork restart, unrestrained resection will lead to fork collapse and genome instability in the absence of fork protection mechanisms (Hashimoto et al., 2010; Schlacher et al., 2011, 2012; Ying et al., 2012; Lemacon et al., 2017; Thangavel et al., 2015; Rondinelli et al., 2017; Liu et al., 2020).
The regulation of post-translational modifications (PTMs) on nucleases at reversed forks could be a critical determinant of sensitivity in cancer treatment. In BRCA1/2 deficient cells, CHD4 and MLL3/4 methylate Histone 3 lysine 4 to recruit MRE11, leading to fork degradation and sensitivity to chemotherapies. Similarly, EZH2 methylates H3K27 to recruit MUS81 to cleave the replication fork, giving rise to chemosensitivity in BRCA1/2 deficient cells. Restoring the fork integrity by either inactivating nucleases MRE11 or MUS81 or depleting the histone methyltransferases leads to RARP inhibitor and other chemotherapy resistance (Ray Chaudhuri et al., 2016; Rondinelli et al., 2017). However, the conflicting evidence from Jasin lab that the separation function mutants of BRCA2 lacking fork protection does not result in the vulnerability of PARP inhibitors and other chemo drugs (Lim et al., 2024). The ssDNA gaps in BRCA2 deficient cells could be also vulnerable to drugs such as PARPi, and nuclease activities are also involved in this process (Cong et al., 2022). In summary, questions such as the replication fork protection, the regulation of nuclease activity by PTMs, and the significance of these regulations for treating BRCA1/2 deficient cancers remain for further study.
UFM1 is a diminutive ubiquitin-like modifier (UBL) that manifests a ubiquitous expression profile across a broad array of eukaryotic organisms. Analogous to the ubiquitin system, UFMylation represents a post-translational modification whereby UFM1 is covalently conjugated to lysine residues on target substrates. This conjugation is facilitated by a concerted enzymatic cascade involving E1 activating enzyme (UBA5), E2 conjugating enzyme (UFC1), and E3 ligase (UFL1) (Peter et al., 2022). The pertinence of UFMylation to human health has been elucidated by the identification of hypomorphic mutations within its machinery, which are implicated in a spectrum of pathologies encompassing cerebellar ataxia, neurodevelopmental anomalies, and skeletal malformations, possibly due to the misfunction in protein homeostasis (Makhlouf et al., 2024). While the significance of UFMylation in the preservation of genome stability has been partially illuminated in recent studies (Lee et al., 2021), its precise role within the replication fork reversal pathway remains to be comprehensively elucidated.
Two recent studies elucidate the regulatory role of UFMylation in the nascent strand degradation within the fork reversal pathway. Gong et al. describe the requisite UFMylation of PARP1 at lysine 548 for the efficacious activation of CHK1 during replication stress. A deficiency in UFL1 impedes CHK1 activation and curtails nascent DNA degradation under replication stress. UFMylation enhances PARP1’s catalytic activity in vitro and facilitates the recruitment of MRE11 at the stalling replication fork. This process, under the aegis of BRCA1/2, facilitates appropriate end resection, conducive to fork restart when stress is released. Furthermore, PARP1 UFMylation-deficient knock-in mice models demonstrate heightened susceptibility to replication stress, characteristic of anti-cancer treatments. The findings suggest that in cells proficient in BRCA1/2, PARP1 UFMylation undergirds CHK1 activation and replication fork integrity during replication stress. Conversely, in BRCA1/2 deficient cells, this mechanism’s promotion of nascent strand degradation may precipitate cellular demise due to excessive resection. In a related vein, Tian et al. expound upon the UFMylation of PTIP, a constituent of the MLL3/4 methyltransferase complex, as a facilitator for the complex’s assembly, engendering an increase in H3K4me1 and H3K4me3 marks at stalled replication forks and subsequent MRE11 recruitment. The UFMylation of PTIP at lysine 148 positions UFL1 as a crucial arbiter of fork stability and the cellular response to PARP inhibitors in BRCA1/2-deficient cells. Their study delineates how UFMylation orchestrates the epigenetic landscape to recruit MRE11 toward the nascent strand of the reversed fork under duress, particularly in cells lacking functional BRCA1/2 (Fig. 1).
In summary, appropriate end resection of the nascent DNA strand is imperative for the facilitation of the fork reversal pathway. BRCA1/2 safeguard the replication fork and the nascent DNA strand against excessive resection, while nucleases such as MRE11 are orchestrated to the nascent strand to mediate resection, producing single-stranded DNA requisite for checkpoint activation in a balanced mechanism. Both studies illuminate the dual roles of UFMylation in modulating MRE11 recruitment, by targeting PARP1 and changing epigenetic environment, a process pivotal in responding to replication stress. Based on the two studies, several critical questions await further investigation. Firstly, PARP1 functions in replication elongation by sensing incompletely processed replication intermediates (Hanzlikova et al., 2018) and together with TIMELESS and TIPIN to protect the replisome in early S phase from transcription–replication conflicts (Petropoulos et al., 2024). The localization of UFMylated PARP1 is yet to be determined, which can’t rule out the possibility that UFMylation can also promote checkpoint activation during replication uncoupling but before fork reversal. Secondly, as MRE11 is also a UFMylation substrate (Lee et al., 2021), the crosstalk between MRE11 UFMylation and PARP/PTIP UFMylations is waiting to be uncovered. Thirdly, as Tian et al. mentioned, how PTIP UFMylation contributes to the assembly of the MLL3/4–PTIP complex and its subsequent impact on methyltransferase activity are unclear, further investigation is warranted. The findings with subsequent studies will not only underscore the contribution of UFMylation to genome stability in normative cellular conditions but also delineate potential therapeutic strategies for the management of BRCA1/2-deficient tumors.
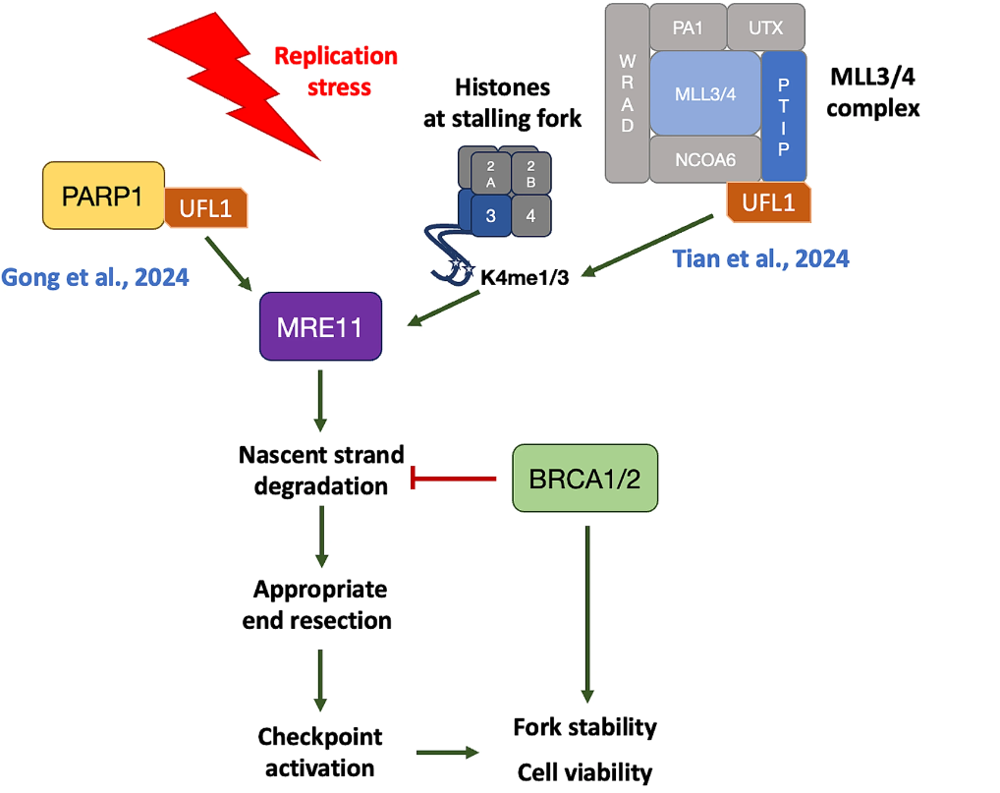 fig 1
fig 1
Data availability
Not applicable.
References
Amunugama, R., Willcox, S., Wu, R. A., Abdullah, U. B., El-Sagheer, A. H., Brown, T., McHugh, P. J., Griffith, J. D., & Walter, J. C. (2018). Replication fork reversal during DNA interstrand Crosslink Repair requires CMG unloading. Cell Rep, 23, 3419–3428. https://doi.org/10.1016/j.celrep.2018.05.061.
Article CAS PubMed PubMed Central Google Scholar
Berti, M., Cortez, D., & Lopes, M. (2020). The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nature Reviews Molecular Cell Biology, 21, 633–651. https://doi.org/10.1038/s41580-020-0257-5.
Article CAS PubMed Google Scholar
Cong, K., & Cantor, S. B. (2022). Exploiting replication gaps for cancer therapy. Molecular Cell, 82, 2363–2369. https://doi.org/10.1016/j.molcel.2022.04.023.
Article CAS PubMed PubMed Central Google Scholar
Cortez, D. (2019). Replication-coupled DNA repair. Molecular Cell, 74, 866–876. https://doi.org/10.1016/j.molcel.2019.04.027.
Article CAS PubMed PubMed Central Google Scholar
Gong, Y., Wang, Z., Zong, W., Shi, R., Sun, W., Wang, S., Peng, B., Takeda, S., Wang, Z. Q., & Xu, X. (2024). PARP1 UFMylation ensures the stability of stalled replication forks. Proc Natl Acad Sci U S A, 121, e2322520121.
Article CAS PubMed PubMed Central Google Scholar
Hanzlikova, H., Kalasova, I., Demin, A. A., Pennicott, L. E., Cihlarova, Z., & Caldecott, K. W. (2018). The importance of poly(ADP-Ribose) polymerase as a Sensor of Unligated Okazaki fragments during DNA replication. Molecular Cell, 71, 319–331e313. https://doi.org/10.1016/j.molcel.2018.06.004.
Article CAS PubMed PubMed Central Google Scholar
Hashimoto, Y., Ray Chaudhuri, A., Lopes, M., & Costanzo, V. (2010). Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nature Structural & Molecular Biology, 17, 1305–1311. https://doi.org/10.1038/nsmb.1927.
Article CAS Google Scholar
Kavlashvili, T., Liu, W., Mohamed, T. M., Cortez, D., & Dewar, J. M. (2023). Replication fork uncoupling causes nascent strand degradation and fork reversal. Nature Structural & Molecular Biology, 30, 115–124. https://doi.org/10.1038/s41594-022-00871-y.
Article CAS Google Scholar
Lee, L., Perez Oliva, A. B., Martinez-Balsalobre, E., Churikov, D., Peter, J., Rahmouni, D., Audoly, G., Azzoni, V., Audebert, S., Camoin, L., et al. (2021). UFMylation of MRE11 is essential for telomere length maintenance and hematopoietic stem cell survival. Science Advances, 7, eabc7371. https://doi.org/10.1126/sciadv.abc7371.
Article CAS PubMed PubMed Central Google Scholar
Lemacon, D., Jackson, J., Quinet, A., Brickner, J. R., Li, S., Yazinski, S., You, Z., Ira, G., Zou, L., Mosammaparast, N., & Vindigni, A. (2017). MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nature Communications, 8, 860. https://doi.org/10.1038/s41467-017-01180-5.
Article CAS PubMed PubMed Central Google Scholar
Lim, P. X., Zaman, M., Feng, W., & Jasin, M. (2024). BRCA2 promotes genomic integrity and therapy resistance primarily through its role in homology-directed repair. Molecular Cell, 84, 447–462e410. https://doi.org/10.1016/j.molcel.2023.12.025.
Article CAS PubMed Google Scholar
Liu, W., Krishnamoorthy, A., Zhao, R., & Cortez, D. (2020). Two replication fork remodeling pathways generate nuclease substrates for distinct fork protection factors. Science Advances, 6. https://doi.org/10.1126/sciadv.abc3598.
Liu, W., Saito, Y., Jackson, J., Bhowmick, R., Kanemaki, M. T., Vindigni, A., & Cortez, D. (2023). RAD51 bypasses the CMG helicase to promote replication fork reversal. Science, 380, 382–387. https://doi.org/10.1126/science.add7328.
Article CAS PubMed PubMed Central Google Scholar
Lopes, M., Foiani, M., & Sogo, J. M. (2006). Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Molecular Cell, 21, 15–27. https://doi.org/10.1016/j.molcel.2005.11.015.
Article CAS PubMed Google Scholar
Makhlouf, L., Peter, J. J., Magnussen, H. M., Thakur, R., Millrine, D., Minshull, T. C., Harrison, G., Varghese, J., Lamoliatte, F., Foglizzo, M., et al. (2024). The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons. Nature, 627, 437–444. https://doi.org/10.1038/s41586-024-07093-w.
Article CAS PubMed PubMed Central Google Scholar
Peter, J. J., Magnussen, H. M., DaRosa, P. A., Millrine, D., Matthews, S. P., Lamoliatte, F., Sundaramoorthy, R., Kopito, R. R., & Kulathu, Y. (2022). A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation. Embo Journal, 41, e111015. https://doi.org/10.15252/embj.2022111015.
Article CAS PubMed PubMed Central Google Scholar
Petropoulos, M., Karamichali, A., Rossetti, G. G., Freudenmann, A., Iacovino, L. G., Dionellis, V. S., Sotiriou, S. K., & Halazonetis, T. D. (2024). Transcription-replication conflicts underlie sensitivity to PARP inhibitors. Nature, 628, 433–441. https://doi.org/10.1038/s41586-024-07217-2.
Article CAS PubMed PubMed Central Google Scholar
Ray Chaudhuri, A., Callen, E., Ding, X., Gogola, E., Duarte, A. A., Lee, J. E., Wong, N., Lafarga, V., Calvo, J. A., Panzarino, N. J., et al. (2016). Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature, 535, 382–387. https://doi.org/10.1038/nature18325.
Article CAS PubMed Google Scholar
Rondinelli, B., Gogola, E., Yucel, H., Duarte, A. A., van de Ven, M., van der Sluijs, R., Konstantinopoulos, P. A., Jonkers, J., Ceccaldi, R., Rottenberg, S., & D’Andrea, A. D. (2017). EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nature Cell Biology, 19, 1371–1378. https://doi.org/10.1038/ncb3626.
Article CAS PubMed Google Scholar
Saldivar, J. C., Cortez, D., & Cimprich, K. A. (2017). The essential kinase ATR: Ensuring faithful duplication of a challenging genome. Nature Reviews Molecular Cell Biology, 18, 622–636. https://doi.org/10.1038/nrm.2017.67.
Article CAS PubMed PubMed Central Google Scholar
Schlacher, K., Christ, N., Siaud, N., Egashira, A., Wu, H., & Jasin, M. (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell, 145, 529–542. https://doi.org/10.1016/j.cell.2011.03.041.
Article CAS PubMed PubMed Central Google Scholar
Schlacher, K., Wu, H., & Jasin, M. (2012). A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell, 22, 106–116. https://doi.org/10.1016/j.ccr.2012.05.015.
Article CAS PubMed PubMed Central Google Scholar
Thakar, T., & Moldovan, G. L. (2021). The emerging determinants of replication fork stability. Nucleic Acids Research, 49, 7224–7238. https://doi.org/10.1093/nar/gkab344.
Article CAS PubMed PubMed Central Google Scholar
Thangavel, S., Berti, M., Levikova, M., Pinto, C., Gomathinayagam, S., Vujanovic, M., Zellweger, R., Moore, H., Lee, E. H., Hendrickson, E. A., et al. (2015). DNA2 drives processing and restart of reversed replication forks in human cells. Journal of Cell Biology, 208, 545–562. https://doi.org/10.1083/jcb.201406100.
Article CAS PubMed PubMed Central Google Scholar
Tian, T., Chen, J., Zhao, H., Li, Y., Xia, F., Huang, J., Han, J., & Liu, T. (2024). UFL1 triggers replication fork degradation by MRE11 in BRCA1/2-deficient cells. Nature Chemical Biology. https://doi.org/10.1038/s41589-024-01611-7.
Article PubMed Google Scholar
Ying, S., Hamdy, F. C., & Helleday, T. (2012). Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Research, 72, 2814–2821. https://doi.org/10.1158/0008-5472.CAN-11-3417.
Article CAS PubMed Google Scholar
Zellweger, R., Dalcher, D., Mutreja, K., Berti, M., Schmid, J. A., Herrador, R., Vindigni, A., & Lopes, M. (2015). Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. Journal of Cell Biology, 208, 563–579. https://doi.org/10.1083/jcb.201406099.
Article CAS PubMed PubMed Central Google Scholar
Download references
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 32161133015 to H.L.); the National Key R&D Program of China (Grant No. 2019YFA0903900 to H.L.); Natural science foundation of Guangdong province of China [Grant No. 2022A1515012495 to H.L.]; SZU Top Ranking Project [Grant No. 86000000210 to H.L.]
Author information
Authors and Affiliations
South China Hospital, Medical School, Guangdong Key Laboratory for Genome Stability & Disease Prevention, Shenzhen University, Shenzhen, 518060, China
Yisui Xia & Huiqiang Lou
Department of Radiation Oncology, University of Virginia School of Medicine, Charlottesville, VA, 22908, USA
Wenpeng Liu
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest and no competing financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Cite this article
Xia, Y., Liu, W. & Lou, H. Dual roles of UFMylation on stalling fork stability. GENOME INSTAB. DIS. 5, 127–130 (2024). https://doi.org/10.1007/s42764-024-00129-5
Download citation
Received
Revised
Accepted
Published
Issue Date
DOIhttps://doi.org/10.1007/s42764-024-00129-5
Share this article
Anyone you share the following link with will be able to read this content:
Provided by the Springer Nature SharedIt content-sharing initiative
Keyword:
DNA replication
Replication stress
Fork stability
Genome stability
Nuclease
Post-translational modifications








用户登录
还没有账号?
立即注册