Diffuse subtype-specific gastric carcinogenesis associated with dysregulation of Notch signaling pathways
Original Research Paper
Published: 14 June 2024
Karthik Balakrishnan
Volume 5, pages 116–126, (2024)
Abstract
Gastric tumors are the third leading cause of cancer-related mortality worldwide. This study investigates the effects of Notch signaling pathway dysregulations during gastric carcinogenesis. Hence, the signatures relevant to Notch signaling pathways were collected from the molecular signatures database, and their expression patterns in available mRNA expression profiles of gastric cancer cohorts were explored using a Z score-based pathway activation scoring method. The results of this study uncover that the Notch signaling pathway signatures are dysregulated highly in diffuse subtype-specific gastric cancer rather than intestinal subtype tumors. In addition, ontological functional analysis reveals that Notch signaling involves extracellular matrix structure and complex organization, collagen structure and trimer biosynthesis during their proliferation, survival, and metastasis. The identified pathway dysregulation is further reconfirmed by examining ROC curves of Notch receptor isoform genes such as NOTCH1, NOTCH2, NOTCH3, and NOTCH4, which could prognostic the diffuse subtype-specific gastric tumors with better specificity and sensitivity with greater areas under the curve (AUC) values. Additionally, overall survival (OS) studies also reassured that these gene expressions reveal poor survival patterns associated with highly expressed conditions in diffuse subtypes of gastric cancer patients with significant p-values (p < 0.05). The results of the genomics drug sensitivity in cancer (GDSC) profile show that ERK and MAPK inhibitors are prominent drug targets for this signaling pathway dysregulations in the corresponding subtype of gastric tumors. Thus, the current findings would benefit the development of drug treatments in diffuse subtype-specific gastric carcinogenesis.
Introduction
Cancers are the second rank in the cause of mortality among non-communicable diseases worldwide (Siegel et al., 2024). Gastric tumors are the third-leading cause of cancer-related demise globally (Iwu & Iwu-Jaja, 2023). The Notch signaling pathways are quietly involved in regulating cell fate during development and homeostasis and have diverse roles in distinct tissues. This signaling is initiated by ligand-receptor-based binding of the following Delta-like and Jagged ligands to Notch receptor isoforms and followed by proteolytic cleavages at these receptors (Koch et al., 2008). The Notch signaling pathway activates or inhibits the signaling transduction (Zhang et al., 2023). Moreover, dysregulation in this signal transduction dictates the conversion of undifferentiated cells into malignant stages (Akbarzadeh et al., 2020). Apart from the gain of function in Notch mutation, inappropriate dysregulation of wild-type Notch signaling pathways also have been reported in many tumors and acts in multiple roles during carcinogenesis (Majumder et al., 2021; Vinson et al., 2016).
Although the Notch pathways are conserved as evolutionary cascades for embryonic and perinatal development. However, the abnormal Notch signaling pathway is also implicated in many cancers’ tumorigenesis (Chung et al., 2023). Based on histopathological features, gastric cancer subtypes are classified into intestinal and diffuse (Lauren, 1965). This study investigates the impacts of Notch signaling pathways dysregulation in gastric carcinogenesis with integrative functional genomics strategies. Analysis of the mRNA expression profiles of gastric cancer shows that diffuse subtype gastric tumors are highly dysregulated with Notch signaling pathways during their growth, proliferation, and survival. This prime information may pave the way for the development of drugs for the treatment of these subtype-specific gastric tumor dysregulations.
Materials and methods
Genome-wide expression profiles
This study collected publicly available genome-wide expression profiles from the repository of the gene expression omnibus (GEO) (Barrett et al., 2013). The following stomach cancer subtypes containing mRNA expression profiles GSE15459, GSE26901, GSE62254, GSE22377, GSE35809, and TCGA were used. Additionally, non-cancerous and gastric cancer comprising mRNA expression profiles GSE13911, GSE54129, and gastric cancer cell lines profile GSE22183 were studied. Microarray datasets were collected in the form of raw.CEL files or MAS5.0/RMA normalized file formats were derived from GEO. Whereas raw.CEL files were further normalized with an R-Affy package (Wilson & Miller, 2005). The probes were mapped to unique gene symbols using the appropriate platform annotation data. Gene expression data from several probes were averaged and used in future investigations.
Pathway activation scoring
Identifying and quantifying complex traits based on a single gene expression pattern is often complicated. Instead, groups or clusters of the genes involved in biological processes are classified as signatures or gene sets that are prominent indicators of cellular processes (Freedman et al., 2011). Molecular signatures database (MSigDB)—a resource that provides signatures modified by various biological or experimental conditions (Subramanian et al., 2005). The signatures were obtained from the MSigDB, an extensive repository of gene sets with more than thirty thousand signatures. The public repository has the following nine [(1) Hallmark signatures, (2) positional signatures, (3) curated signatures, (4) regulatory target gene sets, (5) computational gene sets, (6) ontology gene sets, (7) oncogenic signaling pathway signatures, (8) immunologic signatures, and (9) cell type signatures] different type collections of gene sets or signatures. In the current study, 13 Notch signaling pathway signatures were derived from MSigDB and used to calculate their expression patterns in the profiles of gastric cancer. As established earlier and performed in many studies, Z-score-based pathway activation prediction was used (Balakrishnan et al., 2024; Balakrishnan, 2023a, 2023b; Cheadle et al., 2003). The higher the Z-score, the greater the activation of the signatures in the corresponding tumor samples.
Hierarchical clustering and visualization
Hierarchical clustering is the tool used to detect the group of clustered genes that demonstrate similar expression patterns given biological conditions (Tarca et al., 2006). The hierarchical clustering analysis was accomplished using the dChip tool (Li & Wong, 2003). The clustering of samples and genes was visualized using a heat map illustration. Heatmap is the representation of each gene value encoded by various colors. The red color indicates a higher expression, while the green color in the heatmap image showed a lower expression. Black color represents neutral or no expression. The p-value calculated based on the hypergeometric distribution in the dChip was considered sample-specific cluster enrichment (Li & Wong, 2003). Moreover, ontological functional analysis was performed in MSigDB (Subramanian et al., 2005) and DAVID (the database for annotation, visualization, and integrated discovery) tools (Sherman et al., 2022), respectively. In addition, the STRING database was utilized to analyze the protein–protein interactions (https://string-db.org/) (Szklarczyk et al., 2020).
ROC curve
Different types of subtypes are involved in gastric carcinogenesis. The expression values or mean expression values of the Notch receptor isoform genes like NOTCH1, NOTCH2, NOTCH3, and NOTCH4 were provided as input values for the corresponding subtype-specific gastric tumor samples in the mRNA expression profiles GSE62254 and GSE35809. A receiver operating characteristic (ROC) curve plot was performed using the MedCalc software (Balakrishnan, 2022; DeLong et al., 1988).
Survival curve
The survival curve plots are executed by the Kaplan–Meier plot (Balakrishnan, 2024; Győrffy, 2024). The Kaplan–Meier plot sources have follow-up clinical pieces of information and death rates of information. This tool was used to examine the impact of the corresponding gene expression on the survival pattern of stomach cancer patients. Therefore, Notch receptor isoform genes such as NOTCH1, NOTCH2, NOTCH3, and NOTCH4 were used for plot investigation in the diffuse subtype-specific gastric cancer patient cohorts. The log-rank test was used for p-value calculations (Bland & Altman, 2004).
Genomics drug sensitivity in cancer (GDSC) exploration
Expression patterns were examined across the gastric cancer cell lines profile GSE22183 to identify the drugs targeting Notch signaling pathways. The result analysis showed that the gastric cancer cell lines had elevated expression of the Notch gene set. The drug sensitivity existing as IC50 values in the genomics of drug sensitivity in cancer (GDSC) database was explored to the corresponding gastric cancer cell lines (Yang et al., 2013). The drugs that exhibited a negative correlation between their IC50 and the Notch gene sets' expression pattern were short-listed in MS Excel. Then, a bar graph representing this negative correlation was generated.
Results
Notch signaling pathways are predominantly dysregulated in diffuse subtype gastric cancer
In the study exploring the impacts of Notch signaling pathways in stomach carcinogenesis, the signatures related to Notch signaling pathways were collected from the molecular signatures database (MSigDB) (Table S1). In total, 13 signatures were collected and examined in the mRNA expression profiles of gastric tumors GSE15459, GSE26901, and GSE62254. The Z-score pathway activation pattern suggests that these signaling pathway signatures are highly dysregulated in diffuse subtypes of gastric tumor carcinogenesis (Fig. 1A–C). Moreover, the clustered enrichment of subtype-specific p values is statistically significant (p < 0.05). Thus, the study findings show that the diffuse subtype's carcinogenesis is associated with Notch signaling pathways.
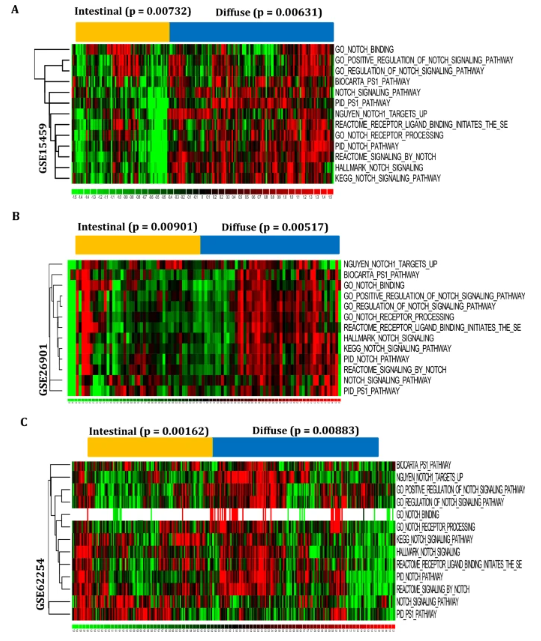 fig 1
fig 1
In the current study, the effects of dysregulations of the Notch signaling pathways during the development of stomach cancer were investigated. A–C) Notch signatures were gathered from the molecular signatures database (MSigDB), and their activation patterns were examined across the mRNA expression profiles of gastric cancer GSE15459 (A), GSE26901 (B), and GSE62254 (C). The Z-score activation pattern indicates that Notch signatures are highly enriched in diffuse subtypes compared to intestinal-type gastric tumors. The p-values for each subtype clustered enrichment of the samples were calculated using the dChip tool's hypergeometric distribution
Notch gene set also highly enriched in diffuse subtype gastric tumors
Apart from signaling pathway signature enrichments, other approaches were employed to identify these pathway dysregulation impacts on diffuse subtype-specific gastric carcinogenesis. Therefore, Notch signaling pathway genes or gene sets that repeatedly occur more than five times across the collected 13 Notch signatures were considered as Notch gene sets and used for further analysis (Table S2). Thus, the derived Notch gene set was studied in normal gastric tissues and gastric cancer subtypes comprising mRNA expression profiles GSE13911, GSE54129, GSE22377, GSE62254, GSE35809, and RNA sequence profile TCGA. The Notch gene set is activated more highly in gastric cancer tissues rather than in normal tissue samples (Fig. 2A, B). Compared to the intestinal subtype, this gene set is strongly activated in diffuse subtype gastric tumors (Fig. 2C, D). Gene-set enrichment analysis (GSEA) of this gene set was also investigated for further confirmation. The GSEA results are also elevated obviously in diffuse subtype-specific gastric cancer with significant enrichment scores (Fig. 2E, F). Thus, these signaling pathway dysregulation patterns have been reconfirmed and are contributing a necessary role in diffuse subtype-specific gastric carcinogenesis.
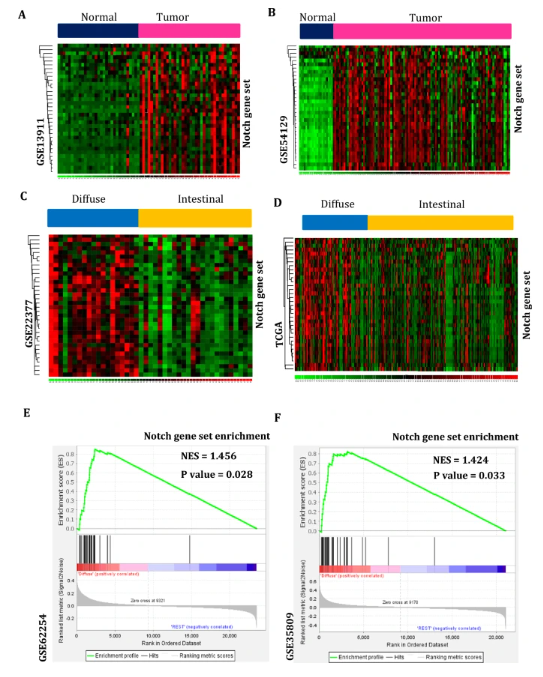 fig 2
fig 2
Additionally, Notch signaling genes or gene sets that repeatedly occur across the collected Notch signatures were explored in non-cancerous gastric tissues and gastric cancer subtypes containing profiles GSE13911, GSE54129, GSE22377, GSE62254, GSE35809, and TCGA. A-B) The Notch gene set is expressed more exclusively in gastric tumor samples than in normal tissues. C-D) This gene set is elevated in diffuse-type gastric tumors compared to the intestinal subtype. In addition, gene-set enrichment analysis (GSEA) of the Notch gene set was also studied for further confirmation. E–F) The GSEA results of this gene set are also enhanced in diffuse subtype gastric tumors with significant enrichment scores
Notch gene set significantly contributes to extracellular matrix dysregulation
Moreover, the Notch gene set was further studied to investigate their functional ontological analysis in the molecular signatures database and DAVID tool. These results exhibit the following cellular processes: extracellular matrix structure and complex organization, collagen structure, and trimer biosynthesis (Fig. 3A, B). The results analysis showed that this Notch signaling pathway involves a crucial role in their proliferation, survival, and metastasis processes. Furthermore, protein–protein interaction was analyzed for a Notch protein in the STRING database. This result also further reconfirmed that the Notch receptor isoform proteins, including Notch-1, Notch-2, Notch-3, and Notch-4, are strongly connected with robust co-expression patterns (Fig. 3C). Thus, all these results clearly show that Notch signaling pathways dysregulation has been supporting cell proliferation, survival, and metastasis processes of the diffuse subtype gastric tumors.
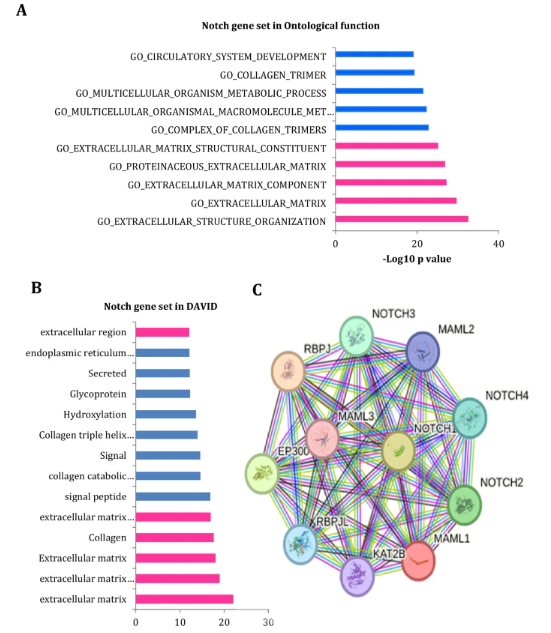 fig 3
fig 3
Furthermore, the ontological functional analysis of the Notch gene set was examined in the molecular signatures database and DAVID tool. A-B) The result shows that the following cellular processes involve extracellular matrix structure and complex organization, including collagen structure and trimer formation. Moreover, the STRING database explored protein–protein interaction for a Notch protein. C) The result also reveals that the Notch receptor isoform proteins are closely associated with higher co-expressions
Notch receptor isoform gene expressions are associated with poor survival in diffuse subtype of gastric cancer patients
Another validation method, a receiver operating characteristic (ROC) curve-based examination, was also achieved for the dysregulation of these pathways in the mRNA expression profiles GSE22377 and GSE15459. The results of the ROC curve for Notch receptor isoform genes such as NOTCH1, NOTCH2, NOTCH3, and NOTCH4 could predict the diffuse subtype-specific gastric tumors with greater specificity and sensitivity with significant areas under the curve (AUC) values (Fig. 4A, B). The results imply that these genes might be contributes to poor prognosis of the diffuse subtype-specific gastric tumors. The impact of Notch receptor isoform gene expressions on the gastric cancer patient's lifespan was also investigated by the overall survival (OS) probability in the diffuse subtypes of patients that were explored in the Kaplan–Meier (KM) plots. The overall survival curve plots of these genes (NOTCH1, NOTCH2, NOTCH3, and NOTCH4) expression show poor survival patterns associated with higher elevated conditions of diffuse subtype gastric cancer patients with significant p-values (p < 0.05) (Fig. 4C–F). Thus, OS curve plots also add extra pieces of evidence that these gene dysregulations are affecting the survival rate of diffuse subtype gastric cancer patients' lifespans.
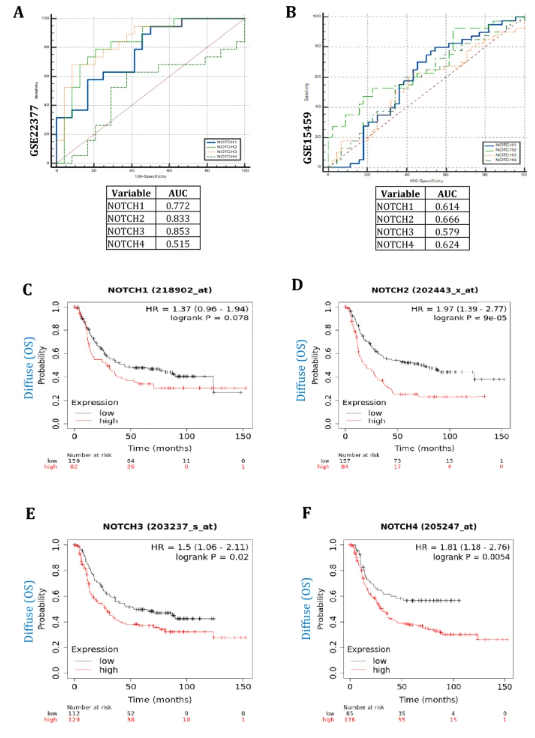 fig4
fig4
The ROC curve-based investigation was also accomplished for the Notch signaling pathways' dysregulation in the profiles GSE22377 and GSE15459 for additional confirmation. A-B) ROC curve of Notch receptors isoform genes such as NOTCH1, NOTCH2, NOTCH3, and NOTCH4 can predict the diffuse subtype with higher specificity and sensitivity. The significant areas under the curve (AUC) values are shown for these genes. Moreover, the effect of these gene expressions on the gastric cancer patient's lifespan was explored. C-F) Hence, the overall survival (OS) probability in the patients with the diffuse subtypes was studied in the Kaplan–Meier (KM) plots. The OS plots of these gene expressions reveal a poor survival pattern accompanied by higher expressed conditions in diffuse subtype patients. The log-rank test was employed to calculate the significant p-values (p < 0.05)
Drugs negatively correlated to Notch signaling pathways
Finally, the genomics drug sensitivity in cancer (GDSC) profile was used to explore the promising drug candidates that target the dysregulated Notch signaling pathways in the corresponding gastric cancer cell line profile GSE22183. The expression patterns of the Notch gene set in the gastric cancer cell line profile are displayed (Fig. 5A). The drugs that show a negative correlation between their IC50 and the expression of the Notch gene set were shown. The drug sensitivity patterns of gastric cancer cell lines to FR-180204, THZ-2-49, Y-39983, TL-1-85, and TPCA-1 were shown in the same order as the expression of the Notch gene set (Fig. 5B–F). There is a negative trend connection between the expression of this gene set and these drugs. The results show the target pathways that correlate the drug with high correlation scores that show a strong negative relationship with the expression of these gene sets (Fig. 5G). ERK and MAPK pathways are notable drug targets on the list that are relevant to the current finding. These drugs may be suitable candidates to target this signaling pathway in a gastric tumor of the diffuse subtype. Thus, the study identifies suitable drug candidates for treating these signaling pathway dysregulations in subtype-specific gastric carcinogenesis. However, it may be necessary to further investigation for more conclusions.
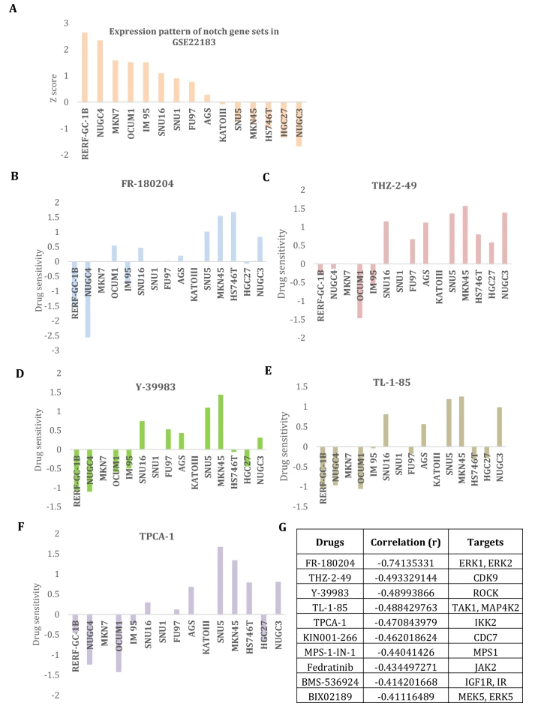 fig 5
fig 5
To investigate the possible therapeutic candidates targeting the dysregulated Notch signaling pathways in gastric cancer cell lines by analysis of genomics drug sensitivity in cancer (GDSC) profile. A) The Notch gene set expression patterns across the gastric cancer cell line profile GSE22183 are shown. B–F) The drugs that exhibit a negative association between their IC50 and the Notch gene set's expression were demonstrated. The sensitivity patterns of gastric cancer cell lines to the following drugs FR-180204 (B), THZ-2–49 (C), Y-39983 (D), TL-1–85 (E), and TPCA-1 (F) presented in the same order as Notch gene set expression in Fig. A. These drugs exhibit a negative trend association with this gene set expression. G) The target pathways corresponding to the drug with correlation higher scores are displayed
Discussion
In animal systems, Notch signaling pathways are well-conserved signaling pathways that determine many critical roles, including homeostasis, differentiation, stem-cell maintenance, and diseases (Siebel & Lendahl, 2017). The Notch pathways are linear-based signaling architecture systems. In a nutshell, Notch receptors are heterodimer structures made with an extracellular domain and transmembrane subunits. Subsequently, it interacted with ligands of transmembrane existing on the nearby cells, leading to receptor subunits segregated by proteolytic cleavage, releasing a Notch intracellular domain (NICD), later transported to the nucleus for the transcriptional regulatory functions (Xia et al., 2022). Thus, Notch receptors and NICD were over-expressed in various cancers (Akbarzadeh et al., 2020; Hopfer et al., 2005; Park et al., 2008). Depending upon the tissue and cellular localization, dysregulated Notch signaling has played complex carcinogenic or tumor-suppressor roles in tissues (Xiu et al., 2020). This study investigates the Notch signaling pathways' impacts on gastric carcinogenesis. The current results suggest that Notch signaling pathways are primarily dysregulated and implicated in diffuse subtype-specific gastric carcinogenesis. Thus, Notch signaling pathway dysregulation plays crucial roles in the various aspects of gastric carcinogenesis including proliferation, growth, survival, and metastasis.
Remarkably, this signaling also can interact with other oncogenic signaling pathways to support tumorigenesis and its progression. For instance, Notch signaling might interact with the NF-kB pathway, contributing to the development of many tumors (Guo et al., 2023). Moreover, dysfunction in Notch receptor isoform expressions can predict prognosis and involves chemoresistance and metastasis (Chen et al., 2010; Jung et al., 2010). The present results also indicate that the Notch receptor isoforms (NOTCH-1, NOTCH-2, NOTCH-3, and NOTCH-4) have a better prognosis sensitivity and specificity rate for the diffuse subtype of gastric tumors. In addition, the results of the overall survival curve plot reveal that these receptors also impact the diffuse subtype of gastric cancer patient’s lifespan. Similarly, previous studies also suggest a significant relationship between the over-expression of these receptors and ovarian cancer survival (Jia et al., 2019). Thus, all the results indicate that Notch signaling pathways play prominent roles in diffuse subtype-specific gastric carcinogenesis.
Conclusion
This study identified that Notch signaling pathways are highly dysregulated during diffuse subtype gastric carcinogenesis. Apart from signatures enrichment, these prominent subtype-specific pathway dysregulation patterns were concluded from that (1) Notch gene sets that are significantly activated in the gastric cancer samples and diffuse subtype tumors, respectively, (2) the ROC curve of Notch receptor isoform gene expressions reveal greater sensitivity and specificity in the corresponding subtype, and (3) the overall survival plot results of these gene expressions also accompanying with poor survival in diffuse-type gastric cancer patients. Thus, dysregulation patterns of the Notch signaling pathways have been identified during subtype-specific gastric carcinogenesis.
Data availability
This article and its online supplementary material include all data generated or investigated during this study.
References
Akbarzadeh, M., Akbarzadeh, S., & Majidinia, M. (2020). Targeting Notch signaling pathway as an effective strategy in overcoming drug resistance in ovarian cancer. Pathology, Research and Practice, 216(11), 153158. https://doi.org/10.1016/j.prp.2020.153158
Article CAS PubMed Google Scholar
Balakrishnan, K. (2022). Hepatocellular carcinoma stage: An almost loss of fatty acid metabolism and gain of glucose metabolic pathways dysregulation. Medical Oncology (northwood, London, England), 39(12), 247. https://doi.org/10.1007/s12032-022-01839-0
Article CAS PubMed Google Scholar
Balakrishnan, K. (2023a). Salt-driven chromatin remodeling associated with senescence dysregulation plays a crucial role in the carcinogenesis of gastric cancer subtype. Computational Toxicology, 25, 100262. https://doi.org/10.1016/j.comtox.2023.100262
Article CAS Google Scholar
Balakrishnan, K. (2023b). The hepatocellular carcinoma (HCC) stage carcinogenesis is associated with genomic instability features. Human Gene, 38, 201228. https://doi.org/10.1016/j.humgen.2023.201228
Article CAS Google Scholar
Balakrishnan, K. (2024). Lactate dehydrogenase isoform expressions differing impacts on gastrointestinal carcinogenesis. Human Gene, 39, 201243. https://doi.org/10.1016/j.humgen.2023.201243
Article CAS Google Scholar
Balakrishnan, K., Chen, Y., & Dong, J. (2024). Amplification of Hippo signaling pathway genes is governed and implicated in the serous subtype-specific ovarian carcinogenesis. Cancers, 16, 1781. https://doi.org/10.3390/cancers16091781
Article CAS PubMed PubMed Central Google Scholar
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., Marshall, K. A., Phillippy, K. H., Sherman, P. M., Holko, M., Yefanov, A., Lee, H., Zhang, N., Robertson, C. L., Serova, N., Davis, S., & Soboleva, A. (2013). NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Research, 41(Database issue), D991-995. https://doi.org/10.1093/nar/gks1193
Article CAS PubMed Google Scholar
Bland, J. M., & Altman, D. G. (2004). The Logrank Test. BMJ (clinical Research Ed.), 328(7447), 1073. https://doi.org/10.1136/bmj.328.7447.1073
Article PubMed Google Scholar
Cheadle, C., Vawter, M. P., Freed, W. J., & Becker, K. G. (2003). Analysis of microarray data using Z score transformation. The Journal of Molecular Diagnostics: JMD, 5(2), 73–81.
Article CAS PubMed PubMed Central Google Scholar
Chen, X., Stoeck, A., Lee, S. J., Shih, I.-M., Wang, M. M., & Wang, T.-L. (2010). Jagged1 expression regulated by Notch3 and Wnt/β-catenin signaling pathways in ovarian cancer. Oncotarget, 1(3), 210–218. https://doi.org/10.18632/oncotarget.127
Article PubMed PubMed Central Google Scholar
Chung, W.-C., & Xu, K. (2023). Notch signaling pathway in pancreatic tumorigenesis. Advances in Cancer Research, 159, 1–36. https://doi.org/10.1016/bs.acr.2023.02.001
Article CAS PubMed Google Scholar
DeLong, E. R., DeLong, D. M., & Clarke-Pearson, D. L. (1988). Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics, 44(3), 837–845.
Article CAS PubMed Google Scholar
Freedman, J. A., Tyler, D. S., Nevins, J. R., & Augustine, C. K. (2011). Use of gene expression and pathway signatures to characterize the complexity of human melanoma. The American Journal of Pathology, 178(6), 2513–2522. https://doi.org/10.1016/j.ajpath.2011.02.037
Article CAS PubMed PubMed Central Google Scholar
Guo, M., Niu, Y., Xie, M., Liu, X., & Li, X. (2023). Notch signaling, hypoxia, and cancer. Frontiers in Oncology, 13, 1078768. https://doi.org/10.3389/fonc.2023.1078768
Article CAS PubMed PubMed Central Google Scholar
Győrffy, B. (2024). Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. British Journal of Pharmacology, 181(3), 362–374. https://doi.org/10.1111/bph.16257
Article CAS PubMed Google Scholar
Hopfer, O., Zwahlen, D., Fey, M. F., & Aebi, S. (2005). The Notch pathway in ovarian carcinomas and adenomas. British Journal of Cancer, 93(6), 709–718. https://doi.org/10.1038/sj.bjc.6602719
Article CAS PubMed PubMed Central Google Scholar
Iwu, C. D., & Iwu-Jaja, C. J. (2023). Gastric cancer epidemiology: Current trend and future direction. Hygiene, 3(3), Article 3. https://doi.org/10.3390/hygiene3030019
Article Google Scholar
Jia, D., Underwood, J., Xu, Q., & Xie, Q. (2019). NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 signaling network is associated with ovarian cancer. Oncology Letters, 17(6), 4914–4920. https://doi.org/10.3892/ol.2019.10170
Article CAS PubMed PubMed Central Google Scholar
Jung, S. G., Kwon, Y. D., Song, J. A., Back, M. J., Lee, S. Y., Lee, C., Hwang, Y. Y., & An, H. J. (2010). Prognostic significance of Notch 3 gene expression in ovarian serous carcinoma. Cancer Science, 101(9), 1977–1983. https://doi.org/10.1111/j.1349-7006.2010.01641.x
Article CAS PubMed PubMed Central Google Scholar
Koch, U., Fiorini, E., Benedito, R., Besseyrias, V., Schuster-Gossler, K., Pierres, M., Manley, N. R., Duarte, A., MacDonald, H. R., & Radtke, F. (2008). Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. The Journal of Experimental Medicine, 205(11), 2515–2523.
Article CAS PubMed PubMed Central Google Scholar
Lauren, P. (1965). The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathologica Et Microbiologica Scandinavica, 64, 31–49. https://doi.org/10.1111/apm.1965.64.1.31
Article CAS PubMed Google Scholar
Li, C., & Wong, W. H. (2003). DNA-chip analyzer (dChip). In G. Parmigiani, E. S. Garrett, R. A. Irizarry, & S. L. Zeger (Eds.), The analysis of gene expression data: Methods and software (pp. 120–141). Springer. https://doi.org/10.1007/0-387-21679-0_5
Chapter Google Scholar
Lobry, C., Oh, P., & Aifantis, I. (2011). Oncogenic and tumor suppressor functions of Notch in cancer: It’s NOTCH what you think. The Journal of Experimental Medicine, 208(10), 1931–1935. https://doi.org/10.1084/jem.20111855
Article CAS PubMed PubMed Central Google Scholar
Majumder, S., Crabtree, J. S., Golde, T. E., Minter, L. M., Osborne, B. A., & Miele, L. (2021). Targeting Notch in oncology: The path forward. Nature Reviews. Drug Discovery, 20(2), 125–144. https://doi.org/10.1038/s41573-020-00091-3
Article CAS PubMed Google Scholar
Park, J. T., Shih, I.-M., & Wang, T.-L. (2008). Identification of Pbx1, a potential oncogene, as a Notch3 target gene in ovarian cancer. Cancer Research, 68(21), 8852–8860. https://doi.org/10.1158/0008-5472.CAN-08-0517
Article CAS PubMed PubMed Central Google Scholar
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., Imamichi, T., & Chang, W. (2022). DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Research, 50(W1), W216–W221. https://doi.org/10.1093/nar/gkac194
Article CAS PubMed PubMed Central Google Scholar
Siebel, C., & Lendahl, U. (2017). Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiological Reviews, 97(4), 1235–1294. https://doi.org/10.1152/physrev.00005.2017
Article CAS PubMed Google Scholar
Siegel, R. L., Giaquinto, A. N., & Jemal, A. (2024). Cancer statistics, 2024. CA: A Cancer Journal for Clinicians, 74(1), 12–49. https://doi.org/10.3322/caac.21820
Article PubMed Google Scholar
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S., & Mesirov, J. P. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America, 102(43), 15545–15550. https://doi.org/10.1073/pnas.0506580102
Article CAS PubMed PubMed Central Google Scholar
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., Doncheva, N. T., Legeay, M., Fang, T., Bork, P., Jensen, L. J., & von Mering, C. (2020). The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research, 49(D1), D605–D612. https://doi.org/10.1093/nar/gkaa1074
Article CAS PubMed Central Google Scholar
Tarca, A. L., Romero, R., & Draghici, S. (2006). Analysis of microarray experiments of gene expression profiling. American Journal of Obstetrics and Gynecology, 195(2), 373–388. https://doi.org/10.1016/j.ajog.2006.07.001
Article CAS PubMed PubMed Central Google Scholar
Vinson, K. E., George, D. C., Fender, A. W., Bertrand, F. E., & Sigounas, G. (2016). The Notch pathway in colorectal cancer. International Journal of Cancer, 138(8), 1835–1842. https://doi.org/10.1002/ijc.29800
Article CAS PubMed Google Scholar
Wilson, C. L., & Miller, C. J. (2005). Simpleaffy: A BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics (oxford, England), 21(18), 3683–3685. https://doi.org/10.1093/bioinformatics/bti605
Article CAS PubMed Google Scholar
Xia, R., Xu, M., Yang, J., & Ma, X. (2022). The role of Hedgehog and Notch signaling pathway in cancer. Molecular Biomedicine, 3(1), 44. https://doi.org/10.1186/s43556-022-00099-8
Article PubMed PubMed Central Google Scholar
Xiu, M.-X., Liu, Y.-M., & Kuang, B.-H. (2020). The oncogenic role of Jagged1/Notch signaling in cancer. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 129, 110416. https://doi.org/10.1016/j.biopha.2020.110416
Article CAS Google Scholar
Yang, W., Soares, J., Greninger, P., Edelman, E. J., Lightfoot, H., Forbes, S., Bindal, N., Beare, D., Smith, J. A., Thompson, I. R., Ramaswamy, S., Futreal, P. A., Haber, D. A., Stratton, M. R., Benes, C., McDermott, U., & Garnett, M. J. (2013). Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Research, 41(Database issue), D955-961. https://doi.org/10.1093/nar/gks1111
Article CAS PubMed Google Scholar
Zhang, H., Yang, Y., Li, X., Yuan, X., & Chu, Q. (2023). Targeting the Notch signaling pathway and the Notch ligand, DLL3, in small cell lung cancer. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 159, 114248. https://doi.org/10.1016/j.biopha.2023.114248
Article Google Scholar
Download references
Acknowledgements
I thank Madurai Kamaraj University for their knowledge and made me a good researcher and my special thanks to Bharathidasan University for their opportunities and made me as a CSIR student, and Anna University, Alagappa University and Bharathiar University also acknowledged.
Funding
No funding was received for this study.
Author information
Authors and Affiliations
Department of Biotechnology, Saroj Institute of Technology and Management (SITM), 12th KM Stone, Lucknow–Sultanpur Road, Lucknow, Uttar Pradesh, 226002, India
Karthik Balakrishnan
Contributions
KB designed the work, performed experiments, analyzed data, and wrote the article.
Corresponding author
Ethics declarations
Conflict of interest
The author declares he has no conflict of interest.
Ethical statement
This article contains no studies with human participants or animal subjects performed.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (XLSX 10 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Cite this article
Balakrishnan, K. Diffuse subtype-specific gastric carcinogenesis associated with dysregulation of Notch signaling pathways. GENOME INSTAB. DIS. 5, 116–126 (2024). https://doi.org/10.1007/s42764-024-00130-y
Download citation
Received
Revised
Accepted
Published
Issue Date
DOIhttps://doi.org/10.1007/s42764-024-00130-y
Share this article
Anyone you share the following link with will be able to read this content:
Provided by the Springer Nature SharedIt content-sharing initiative
Keywords
Gastric cancer
Diffuse subtype
Intestinal subtype
Notch signaling








用户登录
还没有账号?
立即注册